UCVM svm1d and elygtl
From SCECpedia
Contents
New Data
Specifications:
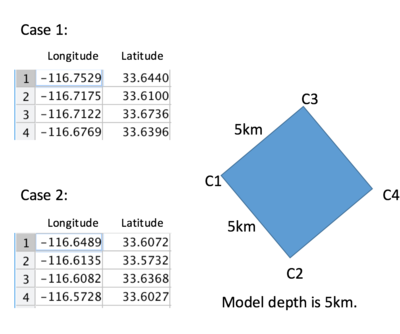
the mpi corner configuration ordering :
mpi-pt0=C1, mpi-pt1=C2, mpi-pt2=C3 and mpi-pt3=C4
with elygtl,
with svm,
etrees,
#!/bin/sh
#Request for etree
#From: Esmaeilzadeh Seylabi, Elnaz <Elnaze@unr.edu>
etree_go () {
sbatch ${SLURM_EXTRACT_SCRIPT}
}
etree_slurm() {
scriptfile=./${SLURM_EXTRACT_SCRIPT}
/bin/cat >$scriptfile << EEND
#!/bin/bash
#SBATCH --ntasks=65
#SBATCH -N 65
#SBATCH --mem 0 # Set to unlimited memory
#SBATCH --time=24:00:00
#SBATCH -o ${PWD}/mpi_${LABEL}_${GTL}_extract_etree.out
#SBATCH -e ${PWD}/mpi_${LABEL}_${GTL}_extract_etree.err
#SBATCH --export=NONE
#SBATCH --mail-user=mei@usc.edu
#SBATCH --mail-type=END
source /home/scec-00/mei/setup_env_proj.sh
###############################################################################
cp ${UCVM_INSTALL}/bin/ucvm2etree_extract_MPI .
cp ${UCVM_INSTALL}/conf/ucvm.conf .
srun -v --mpi=pmi2 ./ucvm2etree_extract_MPI -f ${PWD}/${ETREE_CONFIG}
sbatch ${SLURM_SORT_SCRIPT}
EEND
scriptfile=./${SLURM_SORT_SCRIPT}
/bin/cat >$scriptfile << EEEND
#!/bin/bash
#SBATCH --ntasks=64
#SBATCH -N 64
#SBATCH --mem 0 # Set to unlimited memory
#SBATCH --time=22:00:00
#SBATCH -o ${PWD}/mpi_${LABEL}_${GTL}_sort_etree.out
#SBATCH -e ${PWD}/mpi_${LABEL}_${GTL}_sort_etree.err
#SBATCH --export=NONE
#SBATCH --mail-user=mei@usc.edu
#SBATCH --mail-type=END
source /home/scec-00/mei/setup_env_proj.sh
###############################################################################
cp ${UCVM_INSTALL}/bin/ucvm2etree_sort_MPI .
cp ${UCVM_INSTALL}/conf/ucvm.conf .
srun -v --mpi=pmi2 ./ucvm2etree_sort_MPI -f ${PWD}/${ETREE_CONFIG}
sbatch ${SLURM_MERGE_SCRIPT}
EEEND
scriptfile=./${SLURM_MERGE_SCRIPT}
/bin/cat >$scriptfile << EEEEND
#!/bin/bash
#SBATCH --ntasks=64
#SBATCH -N 64
#SBATCH --mem 0 # Set to unlimited memory
#SBATCH --time=22:00:00
#SBATCH -o ${PWD}/mpi_${LABEL}_${GTL}_merge_etree.out
#SBATCH -e ${PWD}/mpi_${LABEL}_${GTL}_merge_etree.err
#SBATCH --export=NONE
#SBATCH --mail-user=mei@usc.edu
#SBATCH --mail-type=END
source /home/scec-00/mei/setup_env_proj.sh
###############################################################################
cp ${UCVM_INSTALL}/bin/ucvm2etree_merge_MPI .
cp ${UCVM_INSTALL}/conf/ucvm.conf .
srun -v --mpi=pmi2 ./ucvm2etree_merge_MPI -f ${PWD}/${ETREE_CONFIG}
EEEEND
}
## generate the etree.conf
etree_foo() {
configfile=./${ETREE_CONFIG}
/bin/cat >$configfile << END
#
# ${DATE}
#
# ${ETREE_CONFIG}
#
proj=geo-bilinear
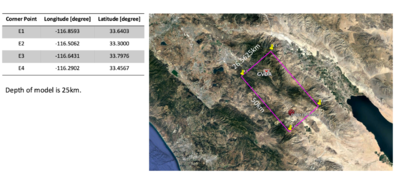
lon_0=-116.8593
lat_0=33.6403
lon_1=-116.6431
lat_1=33.7976
lon_2=-116.2902
lat_2=33.4567
lon_3=-116.5062
lat_3=33.3000
# Domain dimensions (meters):
y-size=26562.5000
x-size=50000.0000
z-size=25000.0000
# Max freq, points per wavelength, Vs min
max_freq=${MAX_FREQ}
ppwl=${PPWL}
vs_min=${VS_MIN}
# Blocks partition parameters:
ny=17
nx=32
# Max allowed size of octants in meters
max_octsize=10000.0
# Etree parameters and info
title=case3_${MAX_FREQ}hz_${PPWL}pts_${VS_MIN}ms
author=Mei_Hui_Su4Elnza
date=09/2019
outputfile=${PWD}/mpi_${LABEL}_${MODEL}_${GTL}_${MAX_FREQ}hz_${PPWL}pts_${VS_MN}ms.e
format=etree
# UCVM parameters
#ucvmstr=${MODEL},svm:svm1d
#ucvm_interp_zrange=Z1
ucvmstr=${MODEL},elygtl:ely
ucvm_interp_zrange=0.0,350.0
ucvmconf=${UCVM_INSTALL}/conf/ucvm.conf
# Scratch
scratch=${PWD}/scratch_case33
#
# Buffering parameters
#
# Etree buffer size in MB
buf_etree_cache=128
# Max octants to buffer for flat file during extraction
buf_extract_mem_max_oct=4194304
# Max octants to save in flat file before reporting full during extraction
buf_extract_ffile_max_oct=40000000
# Max octants to read from input flat file during sorting
buf_sort_ffile_max_oct=40000000
# Minimum number of octants between reports during merging
buf_merge_report_min_oct=10000000
# MPI send/recv octant buffer size during merging
#buf_merge_sendrecv_buf_oct=4096
buf_merge_sendrecv_buf_oct=4096
# Etree read/write octant buffer size during merging
#buf_merge_io_buf_oct=4194304
buf_merge_io_buf_oct=8388608
END
}
##
#label,MODEL,GTL,MAX_FREQ,PPWL,VS_MIN
#
#Elnaz,cvms5,GTL,5,10,200
#Elnaz,cvms5,SVM,5,10,200
#
UCVM_INSTALL="/home/scec-00/mei/ucvmc_19_4/install"
DATE=`date`
input="case_input.txt"
while IFS= read -r file; do
arrData=(${file//,/ })
[[ $arrData =~ ^#.* ]] && continue
LABEL=${arrData[0]}_case33
MODEL=${arrData[1]}
GTL=${arrData[2]}
MAX_FREQ=${arrData[3]}
PPWL=${arrData[4]}
VS_MIN=${arrData[5]}
ETREE_CONFIG=mpi_${LABEL}_${MODEL}_${GTL}_${MAX_FREQ}hz_${PPWL}pts_${VS_MIN}ms.conf
SLURM_EXTRACT_SCRIPT=mpi_${LABEL}_${GTL}_extract_etree.slurm
SLURM_SORT_SCRIPT=mpi_${LABEL}_${GTL}_sort_etree.slurm
SLURM_MERGE_SCRIPT=mpi_${LABEL}_${GTL}_merge_etree.slurm
PWD=`pwd`
etree_foo
etree_slurm
etree_go
done < "$input"
GTL
Exploring UCVMC's external gtl and interp function usage
- elygtl
- svm
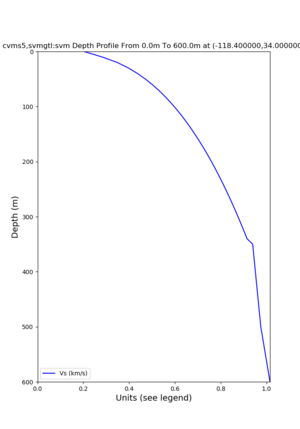
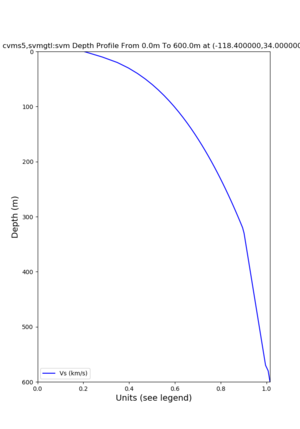
depth profiles
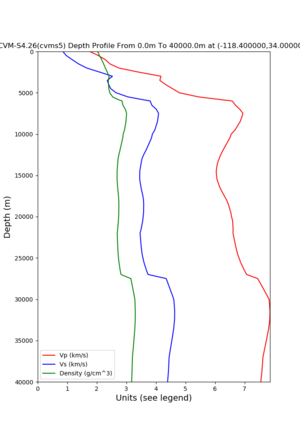
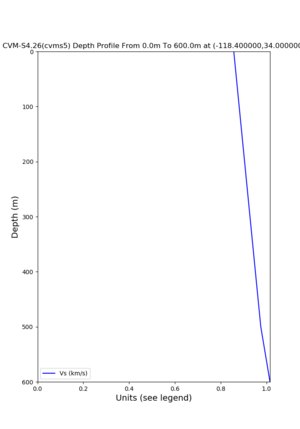
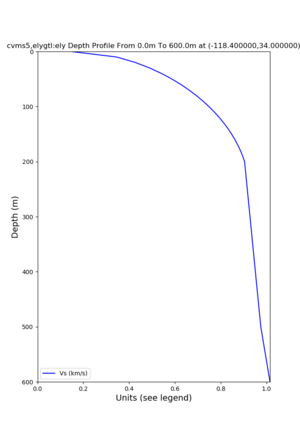
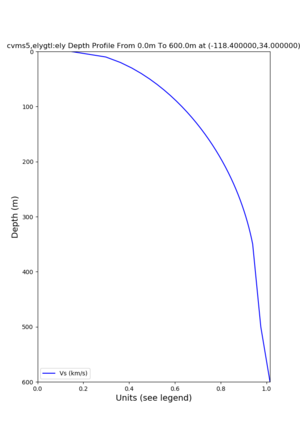
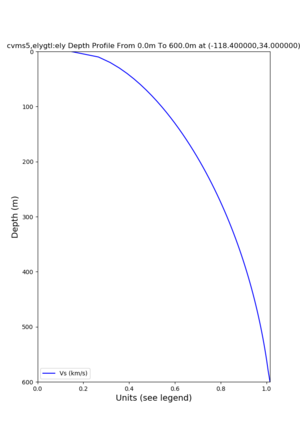
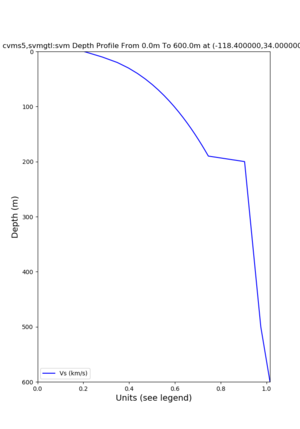
Target point: -118.4,34
echo "-118.4 34.0 " | basin_query -m cvms5 -f ../conf/ucvm.conf
returns the Z1.0 at 580.0
Profile plots,
commands used :
./plot_depth_profile.py -s 34,-118.4 -b 0 -e 40000 -d vs,vp,density -v 100 -c cvms5 -o cvms5_depth_nogtl_bkg.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5 -o cvms5_depth_nogtl_base.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,elygtl:ely -z 0,200 -o cvms5_depth_elygtl_200.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,elygtl:ely -z 0,350 -o cvms5_depth_nogtl_elygtl_350.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,elygtl:ely -Z 1000 -o cvms5_depth_nogtl_elygtl_Z1.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,svmgtl:svm -z 0,200 -o cvms5_depth_nogtl_svmgtl_200.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,svmgtl:svm -z 0,350 -o cvms5_depth_nogtl_svmgtl_350.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,svmgtl:svm -Z 1000 -o cvms5_depth_nogtl_svmgtl_Z1.png
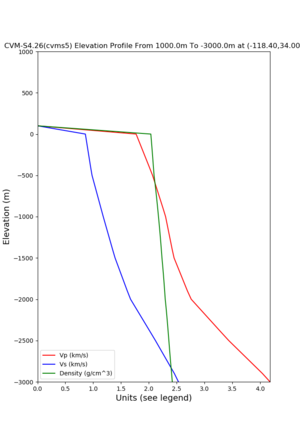
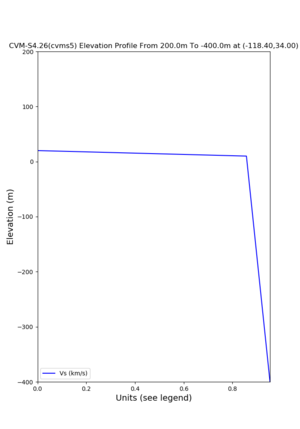
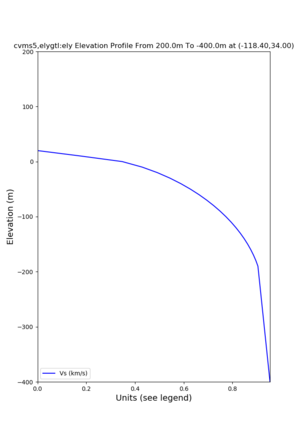
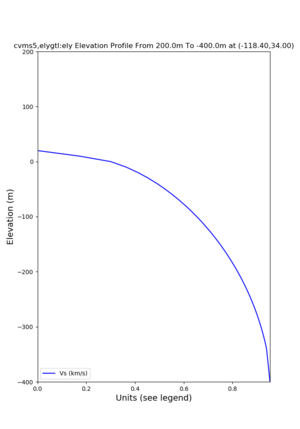
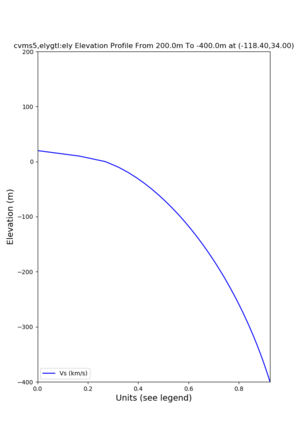
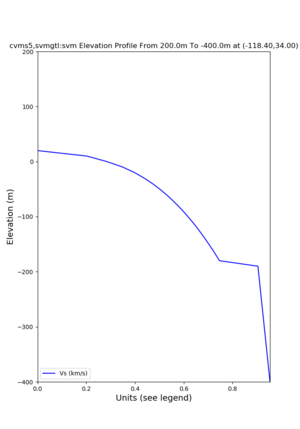
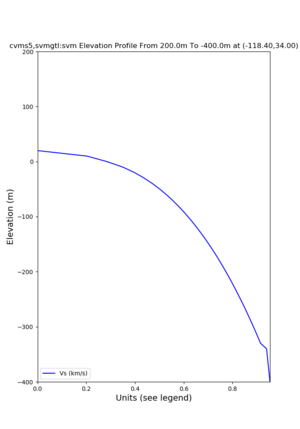
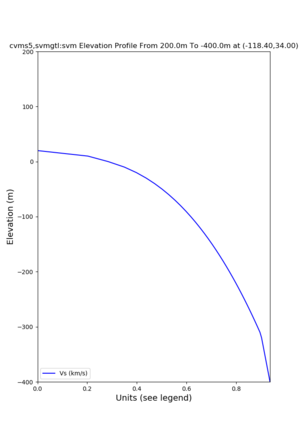
Elevation profile plot,
Commands used:
./plot_elevation_profile.py -s 34,-118.4 -b 1000 -e -3000 -d vs,vp,density -v 100 -c cvms5 -o cvms5_edepth_nogtl_bkg.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5 -o cvms5_edepth_nogtl_base.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,elygtl:ely -z 0,200 -o cvms5_edepth_nogtl_elygtl_200.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,elygtl:ely -z 0,350 -o cvms5_edepth_nogtl_elygtl_350.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,elygtl:ely -Z 1000 -o cvms5_edepth_nogtl_elygtl_Z1.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,svmgtl:svm -z 0,200 -o cvms5_edepth_nogtl_svmgtl_200.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,svmgtl:svm -z 0,350 -o cvms5_edepth_nogtl_svmgtl_350.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,svmgtl:svm -Z 1000 -o cvms5_edepth_nogtl_svmgtl_Z1.png
Somewhere by the water
Near westside with cvms5, internal GTL disabled,
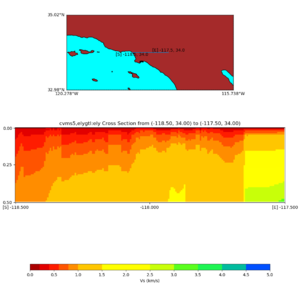
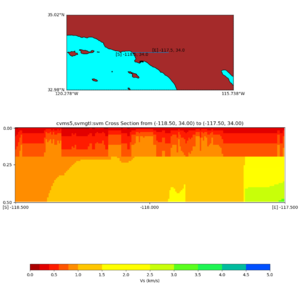
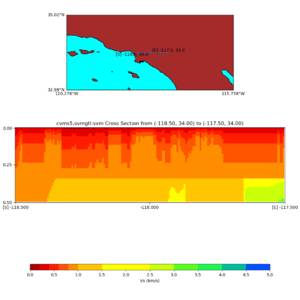
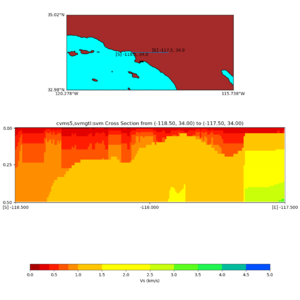
Depth cross plot,
commands used :
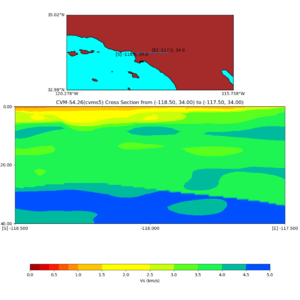
./plot_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 500 -v 10 -d vs -c cvms5 -a d -s 0 -e 500 -o cvms5_cross_nogtl_base.png
and,
./plot_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 500 -v 10 -d vs -c cvms5,svmgtl:svm -a d -Z 1000 -s 0 -e 500 -o cvms5_cross_nogtl_svmgtl_Z1.png
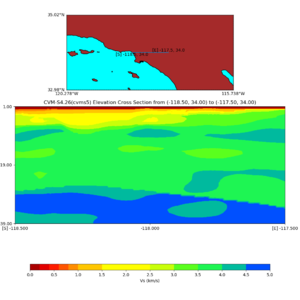
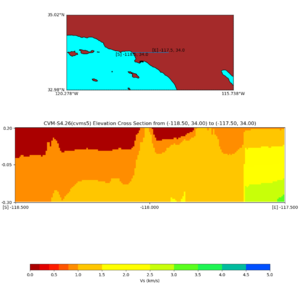
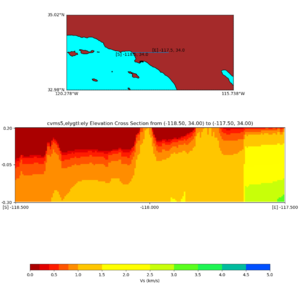
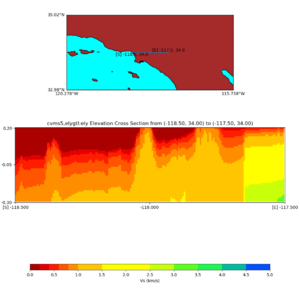
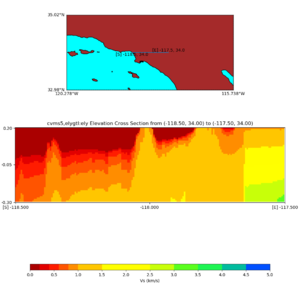
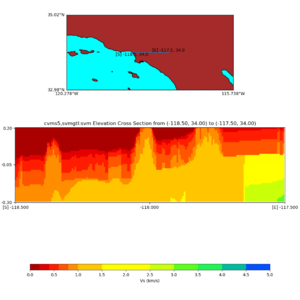
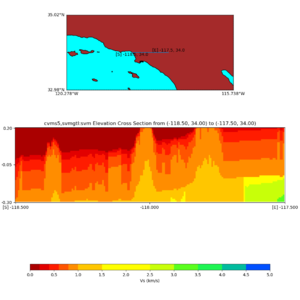
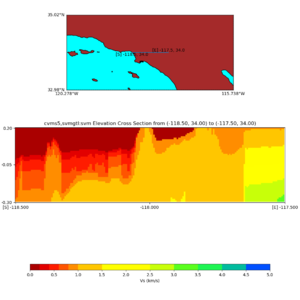
Elevation cross plot, Commands used:
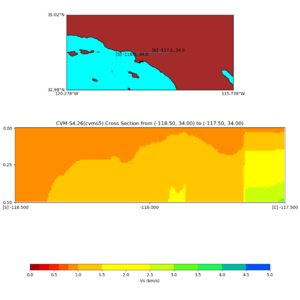
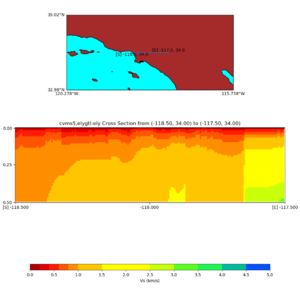
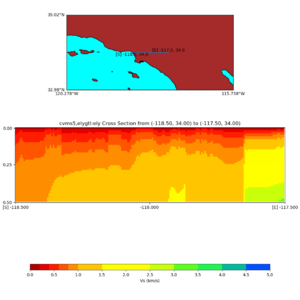
./plot_elevation_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 500 -v -10 -d vs -c cvms5 -a d -s 200 -e -300 -o cvms5_ecross_nogtl_base.png
and,
./plot_elevation_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 500 -v -10 -d vs -c cvms5,svmgtl:svm -a d -Z 1000 -s 200 -e -300 -o cvms5_ecross_nogtl_svmgtl_Z1.png
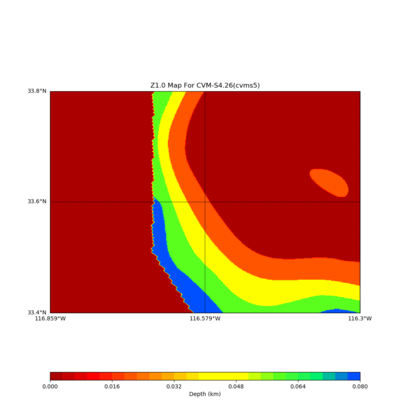
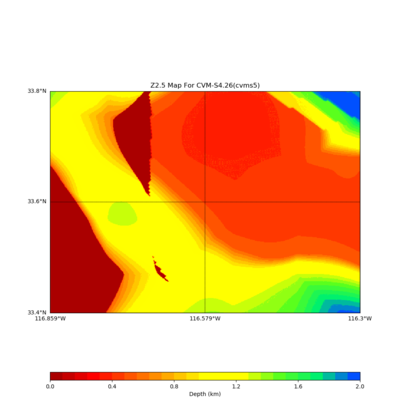
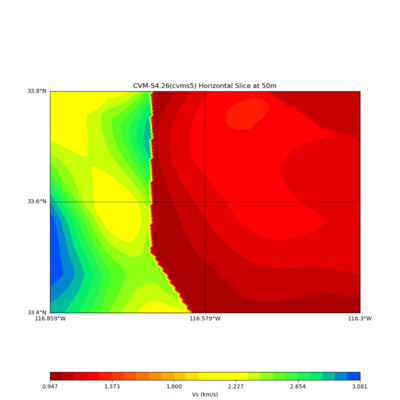
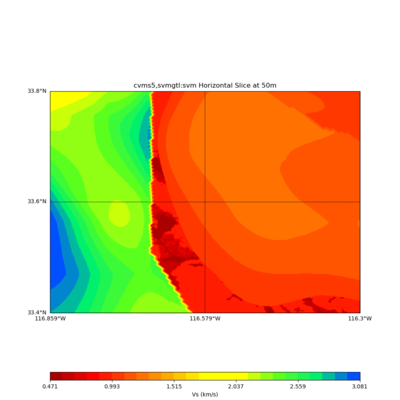
Somewhere near Garner Valley
# gv
LOC='-b 33.40,-116.859 -u 33.80,-116.30'
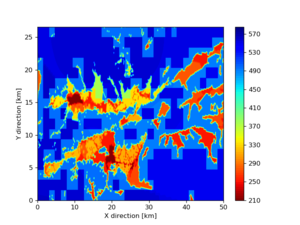
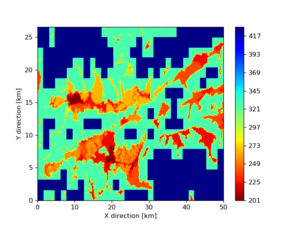
./plot_horizontal_slice.py ${LOC} -e 50 -d vs -c cvms5 -a dd -s 0.001 -o gv_cvms5_vs_50.png
./plot_horizontal_slice.py ${LOC} -e 50 -d vs -c cvms5,svmgtl:svm -Z 1000 -a dd -s 0.001 -o gv_cvms5_svm_vs_50.png
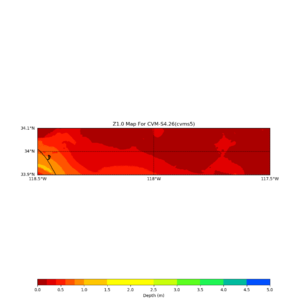
./plot_z10_map.py ${LOC} -c cvms5 -a dd -s 0.001 -o gv_cvms5_Z10.png
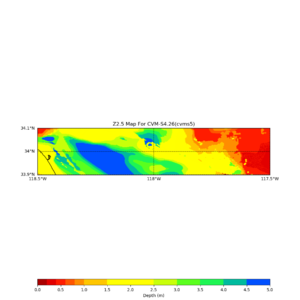
./plot_z25_map.py ${LOC} -c cvms5 -a dd -s 0.001 -o gv_cvms5_Z25.png