Difference between revisions of "UCVM svm1d and elygtl"
From SCECpedia
Jump to navigationJump to search| Line 31: | Line 31: | ||
etrees, | etrees, | ||
| − | [http://hypocenter.usc.edu/research/ucvmc_result/svmgtl/mpi_Elnaz_case33_cvms5_GTL_5hz_10pts_200ms.e case3 with GTL][http://hypocenter.usc.edu/research/ucvmc_result/svmgtl/mpi_Elnaz_case33_cvms5_SVM_5hz_10pts_200ms.e case3 with SVM] | + | * [http://hypocenter.usc.edu/research/ucvmc_result/svmgtl/mpi_Elnaz_case33_cvms5_GTL_5hz_10pts_200ms.e case3 with GTL] |
| + | * [http://hypocenter.usc.edu/research/ucvmc_result/svmgtl/mpi_Elnaz_case33_cvms5_SVM_5hz_10pts_200ms.e case3 with SVM] | ||
== GTL == | == GTL == | ||
Revision as of 04:57, 15 September 2019
Contents
New Data
Specifications:
with elygtl,
with svm,
etrees,
GTL
Exploring UCVMC's external gtl and interp function usage
- elygtl
- svm
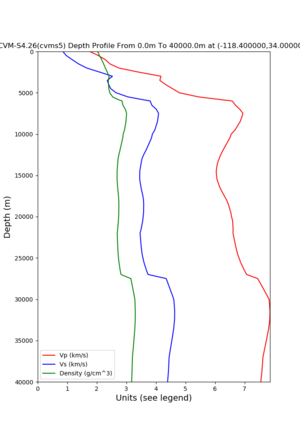
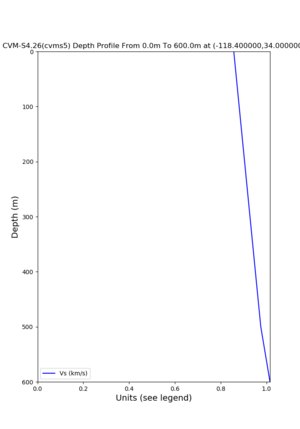
depth profiles
Target point: -118.4,34
echo "-118.4 34.0 " | basin_query -m cvms5 -f ../conf/ucvm.conf
returns the Z1.0 at 580.0
Profile plots,
commands used :
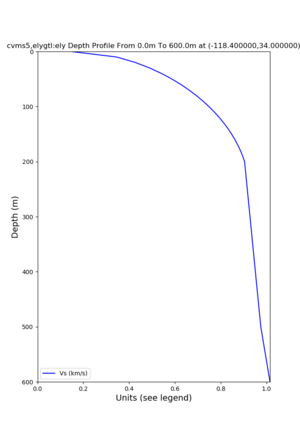
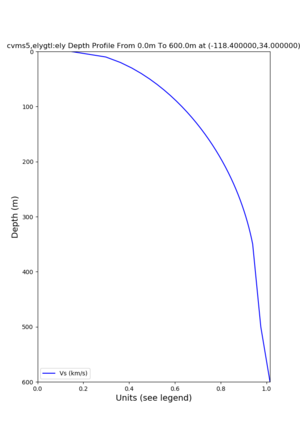
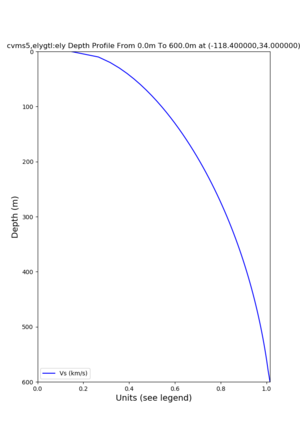
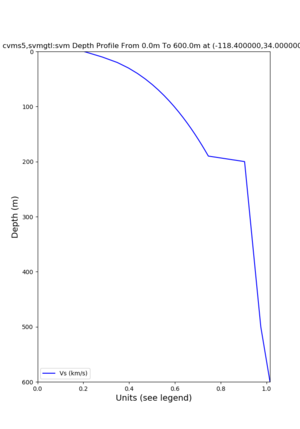
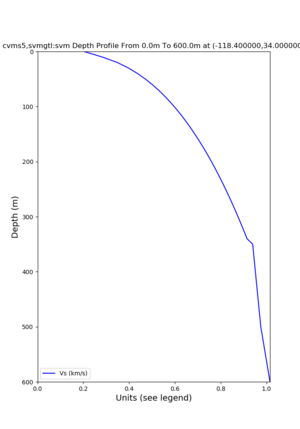
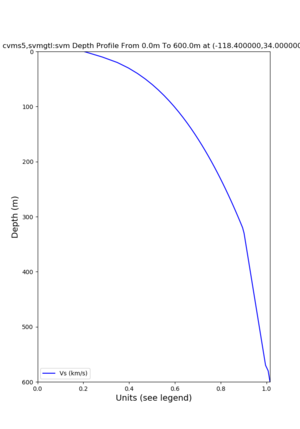
./plot_depth_profile.py -s 34,-118.4 -b 0 -e 40000 -d vs,vp,density -v 100 -c cvms5 -o cvms5_depth_nogtl_bkg.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5 -o cvms5_depth_nogtl_base.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,elygtl:ely -z 0,200 -o cvms5_depth_elygtl_200.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,elygtl:ely -z 0,350 -o cvms5_depth_nogtl_elygtl_350.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,elygtl:ely -Z 1000 -o cvms5_depth_nogtl_elygtl_Z1.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,svmgtl:svm -z 0,200 -o cvms5_depth_nogtl_svmgtl_200.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,svmgtl:svm -z 0,350 -o cvms5_depth_nogtl_svmgtl_350.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 500 -d vs -v 10 -c cvms5,svmgtl:svm -Z 1000 -o cvms5_depth_nogtl_svmgtl_Z1.png
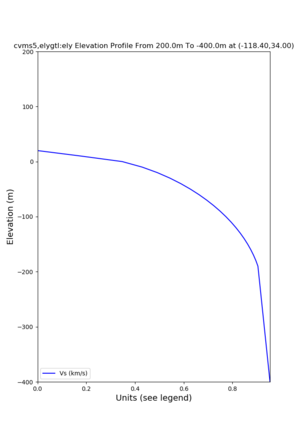
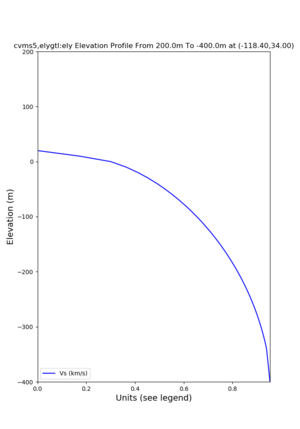
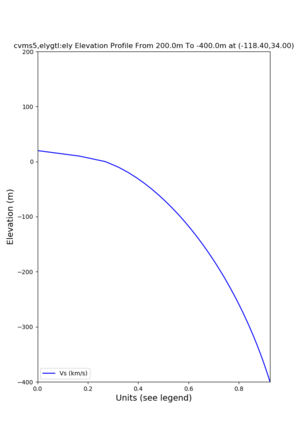
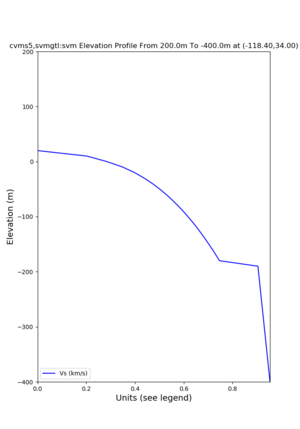
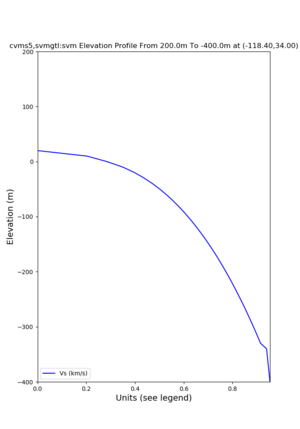
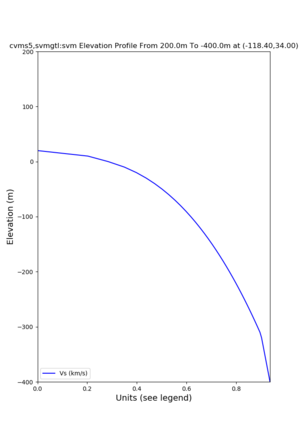
Elevation profile plot,
Commands used:
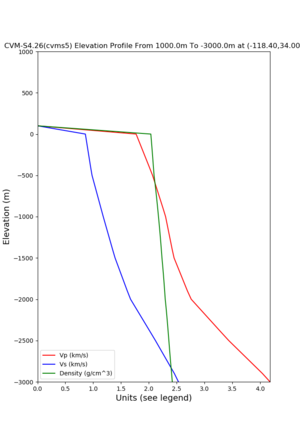
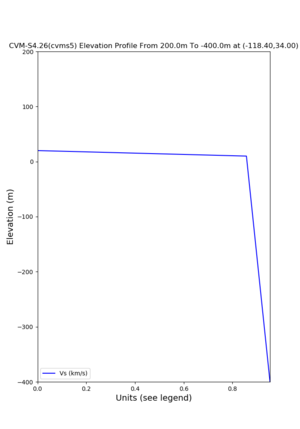
./plot_elevation_profile.py -s 34,-118.4 -b 1000 -e -3000 -d vs,vp,density -v 100 -c cvms5 -o cvms5_edepth_nogtl_bkg.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5 -o cvms5_edepth_nogtl_base.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,elygtl:ely -z 0,200 -o cvms5_edepth_nogtl_elygtl_200.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,elygtl:ely -z 0,350 -o cvms5_edepth_nogtl_elygtl_350.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,elygtl:ely -Z 1000 -o cvms5_edepth_nogtl_elygtl_Z1.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,svmgtl:svm -z 0,200 -o cvms5_edepth_nogtl_svmgtl_200.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,svmgtl:svm -z 0,350 -o cvms5_edepth_nogtl_svmgtl_350.png ./plot_elevation_profile.py -s 34,-118.4 -b 200 -e -300 -d vs -v -10 -c cvms5,svmgtl:svm -Z 1000 -o cvms5_edepth_nogtl_svmgtl_Z1.png
Somewhere by the water
Near westside with cvms5, internal GTL disabled,
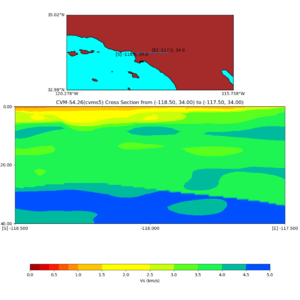
Depth cross plot,
commands used :
./plot_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 500 -v 10 -d vs -c cvms5 -a d -s 0 -e 500 -o cvms5_cross_nogtl_base.png
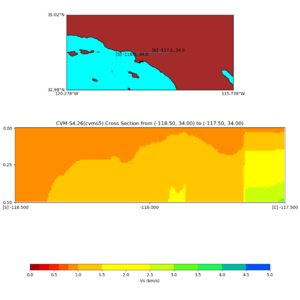
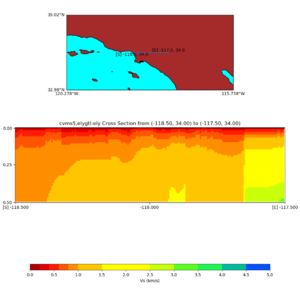
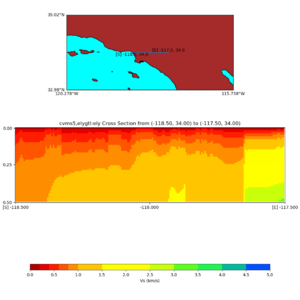
and,
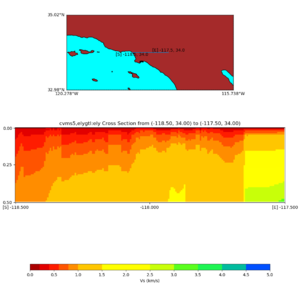
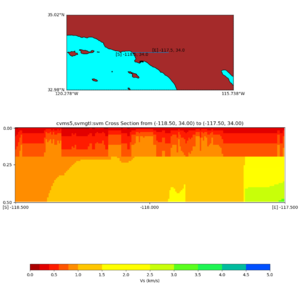
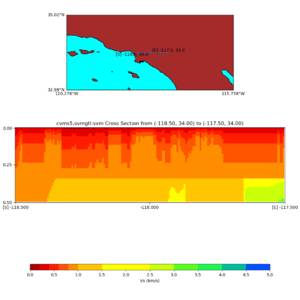
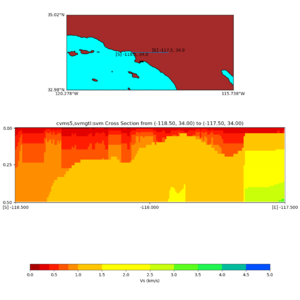
./plot_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 500 -v 10 -d vs -c cvms5,svmgtl:svm -a d -Z 1000 -s 0 -e 500 -o cvms5_cross_nogtl_svmgtl_Z1.png
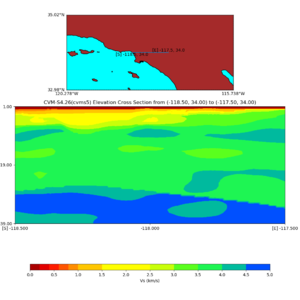
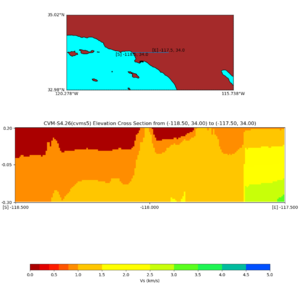
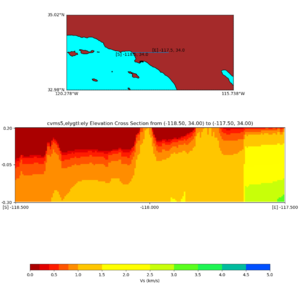
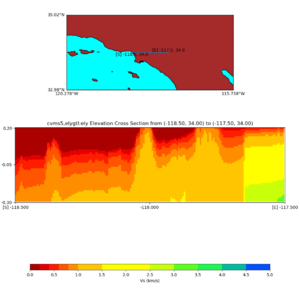
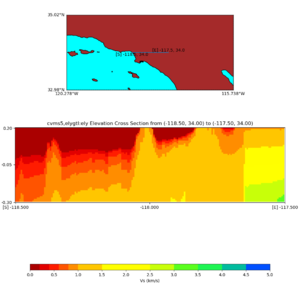
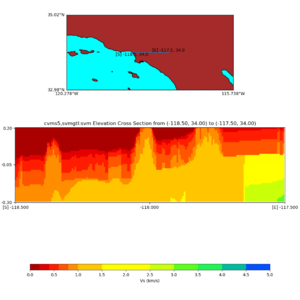
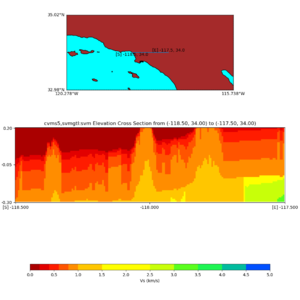
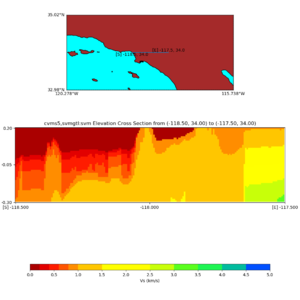
Elevation cross plot, Commands used:
./plot_elevation_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 500 -v -10 -d vs -c cvms5 -a d -s 200 -e -300 -o cvms5_ecross_nogtl_base.png
and,
./plot_elevation_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 500 -v -10 -d vs -c cvms5,svmgtl:svm -a d -Z 1000 -s 200 -e -300 -o cvms5_ecross_nogtl_svmgtl_Z1.png
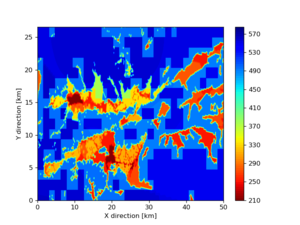
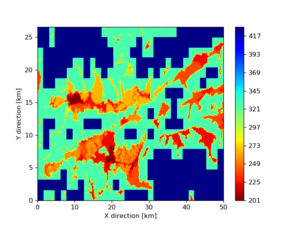
Somewhere near Garner Valley
# gv
LOC='-b 33.40,-116.859 -u 33.80,-116.30'
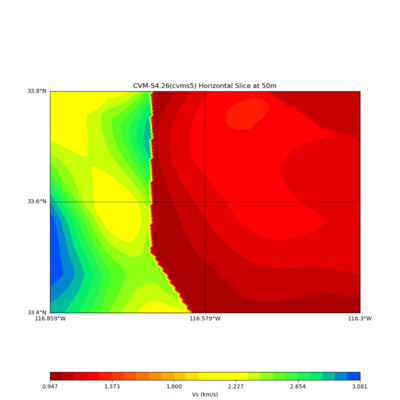
./plot_horizontal_slice.py ${LOC} -e 50 -d vs -c cvms5 -a dd -s 0.001 -o gv_cvms5_vs_50.png
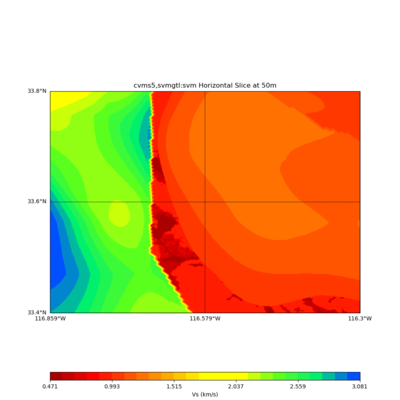
./plot_horizontal_slice.py ${LOC} -e 50 -d vs -c cvms5,svmgtl:svm -Z 1000 -a dd -s 0.001 -o gv_cvms5_svm_vs_50.png
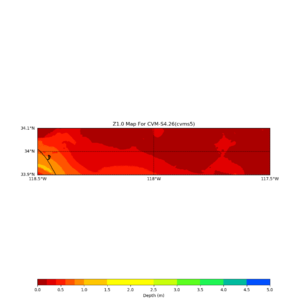
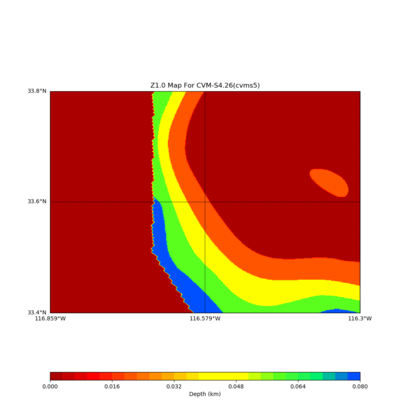
./plot_z10_map.py ${LOC} -c cvms5 -a dd -s 0.001 -o gv_cvms5_Z10.png
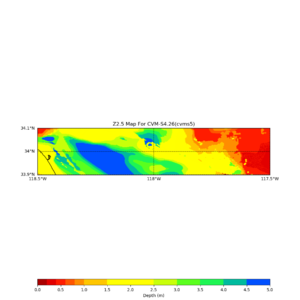
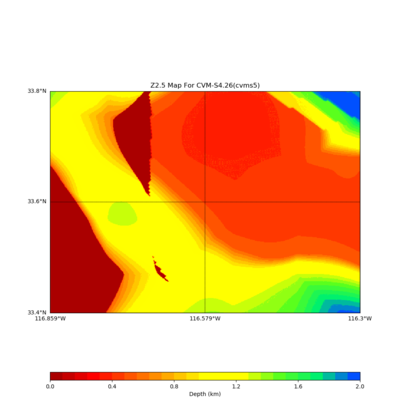
./plot_z25_map.py ${LOC} -c cvms5 -a dd -s 0.001 -o gv_cvms5_Z25.png