Difference between revisions of "NorCal CyberShake SW4 Mesh"
| Line 949: | Line 949: | ||
As a result, I think this is a squashing artifact, which we should report but not necessarily worry about fixing for this study. Please let me know if you feel differently. | As a result, I think this is a squashing artifact, which we should report but not necessarily worry about fixing for this study. Please let me know if you feel differently. | ||
</pre> | </pre> | ||
| + | |||
| + | == additional plots at s3171 == | ||
| + | |||
| + | <pre> | ||
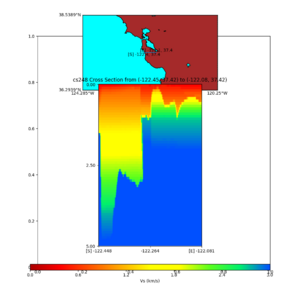
| + | plot_cross_section.py -s 0 -h 204 -d vs -c sfcvm -a sd -o sfcvm_vs_s3171_5k_l_c.png -i $UCVM_INSTALL_PATH -v | ||
| + | 20 -b 37.4164,-122.448 -u 37.4164,-122.081 -e 5000 | ||
| + | plot_cross_section.py -s 0 -h 204 -d vs -c cs248 -a sd -o cs248_vs_s3171_5k_l_c.png -i $UCVM_INSTALL_PATH -v | ||
| + | 20 -b 37.4164,-122.448 -u 37.4164,-122.081 -e 5000 | ||
| + | |||
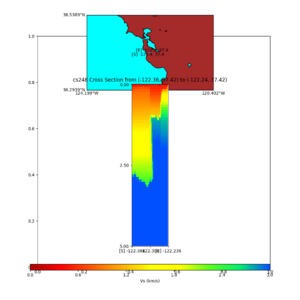
| + | plot_cross_section.py -s 0 -h 204 -d vs -c sfcvm -a sd -o sfcvm_vs_s3171_5k_c.png -i $UCVM_INSTALL_PATH -v 2 | ||
| + | 0 -b 37.4164,-122.3637 -u 37.4164,-122.2359 -e 5000 | ||
| + | plot_cross_section.py -s 0 -h 204 -d vs -c cs248 -a sd -o cs248_vs_s3171_5k_c.png -i $UCVM_INSTALL_PATH -v 2 | ||
| + | 0 -b 37.4164,-122.3637 -u 37.4164,-122.2359 -e 5000 | ||
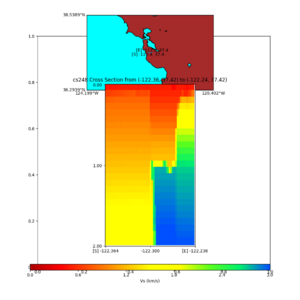
| + | plot_cross_section.py -s 0 -h 204 -d vs -c sfcvm -a sd -o sfcvm_vs_s3171_2k_c.png -i $UCVM_INSTALL_PATH -v 2 | ||
| + | 0 -b 37.4164,-122.3637 -u 37.4164,-122.2359 -e 2000 | ||
| + | plot_cross_section.py -s 0 -h 204 -d vs -c cs248 -a sd -o cs248_vs_s3171_2k_c.png -i $UCVM_INSTALL_PATH -v 2 | ||
| + | 0 -b 37.4164,-122.3637 -u 37.4164,-122.2359 -e 2000 | ||
| + | </pre> | ||
| + | |||
| + | {| | ||
| + | | [[FILE:sfcvm_vs_s3171_5k_l_c.png|thumb|300px|]] | ||
| + | | [[FILE:sfcvm_vs_s3171_5k_c.png|thumb|300px|]] | ||
| + | | [[FILE:sfcvm_vs_s3171_2k_c.png|thumb|300px|]] | ||
| + | |} | ||
| + | |||
| + | {| | ||
| + | | [[FILE:cs248_vs_s3171_5k_l_c.png|thumb|300px|]] | ||
| + | | [[FILE:cs248_vs_s3171_5k_c.png|thumb|300px|]] | ||
| + | | [[FILE:cs248_vs_s3171_2k_c.png|thumb|300px|]] | ||
| + | |} | ||
== Related Entries == | == Related Entries == | ||
*[[UCVM]] | *[[UCVM]] | ||
Revision as of 16:36, 26 August 2024
- Create CyberShake Mesh for NorCal in 3 files (80m)
- Tile with background
Contents
Pre-study
goal: to validate the sw4 plot of sfcvm native hdf5 datafile via its gmg reader shows topography features at surface and yet
accessing UCVM's sfcvm is showing a chopped plot at Z=0
Frontier setup
modules :
module load cray-python module unload PrgEnv-cray module load PrgEnv-gnu gcc module load libtool/2.4.6module load Core/24.07 module load openblas/0.3.26
building UCVM
#!/bin/bash # hn=`hostname -d` ppwd=`pwd` export MY_TOP=$ppwd/scratch export TOP_UCVM_TARGET=$MY_TOP/TARGET_UCVM_SFCVM export UCVM_SRC_PATH=$TOP_UCVM_TARGET/UCVM export UCVM_INSTALL_PATH=$TOP_UCVM_TARGET/ucvm_install rm -rf $TOP_UCVM_TARGET mkdir $TOP_UCVM_TARGET cd $TOP_UCVM_TARGET git clone https://github.com/SCECcode/ucvm.git -b withSFCVM UCVM cd $UCVM_SRC_PATH/largefiles ./get_largefiles.py -m sfcvm,cca,cvmsi cd $UCVM_SRC_PATH/largefiles; ./stage_largefiles.py cd $UCVM_SRC_PATH ./ucvm_setup.py -d -a -p $UCVM_INSTALL_PATH &> ucvm_setup_install.log echo "..EXITING.." exit
SPECIAL NOTE 1:
on Frontier, the first time ucvm_setup.py call seems to fail when configure is called via script. If rerun by hand, It actually goes through.
1) extract the configure command from config.log and run it by hand
2) at UCVM top level, call
make clean; make; make install
3) make sure ucvm_env.sh is setup properly
./ucvm_setup.py -a -d -r -p $UCVM_INSTALL_PATH
4) source $UCVM_INSTALL_PATH/conf/uvcm_env.sh
SPECIAL NOTE 2:
for sw4, hdf5 needs to be enabled with parallel option,
1) cd $UCVM_SRC_PATH/work/lib/hdf5
2) extract the configure command from config.log, and rerun it with --enable-parallel
3) and then call
make clean; make; make install;
building sw4
git clone https://github.com/geodynamics/sw4.git sw4-clone
git clone https://github.com/geodynamics/sw4.git -b ucvm_cmd sw4-ucvm_cmd
building sw4-clone,
add this file, config.inc, under configs directory
FC = gfortran CXX = mpicxx proj=yes hdf5=yes SW4ROOT = /ccs/home/mei/scratch/TARGET_UCVM_SFCVM/ucvm_install/lib/proj SQL3ROOT = /ccs/home/mei/scratch/TARGET_UCVM_SFCVM/ucvm_install/lib/sqlite HDF5ROOT = /ccs/home/mei/scratch/TARGET_UCVM_SFCVM/ucvm_install/lib/hdf5 EXTRA_LINK_FLAGS = /ccs/home/mei/scratch/TARGET_UCVM_SFCVM/ucvm_install/lib/openssl/lib/libcrypto.so.3 /usr/lib64/libopenblas.so.0 -L/opt/cray/pe/gcc/12.2.0/snos/lib64 -lgfortran
top level Makefile needs to be tweaked alittle..
diff --git a/Makefile b/Makefile index 171d1c5a..ca1ef313 100644 --- a/Makefile +++ b/Makefile @@ -78,6 +78,8 @@ SW4LIB64 = $(SW4ROOT)/lib64 #Default, override with configs/make.name. Preferably, FFTW is installed under SW4ROOT FFTWHOME = $(SW4ROOT) +SQL3INC = $(SQL3ROOT)/include +SQL3LIB = $(SQL3ROOT)/lib emptystring := "" foundincfile := $(emptystring) @@ -174,8 +176,8 @@ ifdef EXTRA_FORT_FLAGS endif ifeq ($(proj),yes) - CXXFLAGS += -DENABLE_PROJ -I$(SW4INC) - linklibs += -L$(SW4LIB) -L$(SW4LIB64) -lproj -lsqlite3 -Wl,-rpath,$(SW4LIB) -Wl,-rpath,$(SW4LIB64) + CXXFLAGS += -DENABLE_PROJ -I$(SW4INC) -I$(SQL3INC) + linklibs += -L$(SW4LIB) -L$(SW4LIB64) -lproj -L$(SQL3LIB) -lsqlite3 -Wl,-rpath,$(SW4LIB) -Wl,-rpath,$(SW4LIB64) endif
compile sw4 by
make
making sfcvm mesh in sfile format using sw4
building sw4-ucvm_clone is just the same as sw4-stock but it is alittle ad-hoc and so just assume it is done and the sfcvm_mesh.sfile is here..
ccs/home/mei/proj-geo156/ucvm/sw4/sfcvm.output/sfcvm_mesh.sfile
the sfcvm.txt used for generate this sfcvm_mesh.sfile,
# SW4 SCRIPT fileio path=./sfcvm.output pfs=1 verbose=1 printcycle=10 nwriters=16 time steps=10 # GRID SIZE AND REFINEMENT grid x=280000 y=115000 z=30000 h=100.0 lat=39.111563 lon=-122.569504 az=143.638 proj=tmerc datum=NAD83 lon_p=-123.0 lat_p=35.0 scale=0. 9996 supergrid gp=30 time t=0.1 globalmaterial vsmin=400 vpmin=800 attenuation phasefreq=1.0 nmech=3 maxfreq=10.0 ucvm filename=sfcvm.h100 # POINT SOURCE source depth=8320 lat=36.86733 lon=-121.6105 m0=2.515e15 dip=80 strike=138 rake=-177 t0=1.0 type=Dirac # Station rec lat=37.52886 lon=-122.25522 depth=0 file=1002_37.52886_-122.25522 variables=velocity nsew=1 usgsformat=1 sacformat=0 sfileoutput file=sfcvm.mesh
and the slurm script used is,
#!/bin/bash #SBATCH -J sfcvm_sw4 # Job name #SBATCH -o sfcvm_sw4.oo # Name of stdout output file #SBATCH -e sfcvm_sw4.ee # Name of stderr error file #SBATCH -N 5 # Total # of nodes #SBATCH -n 100 #SBATCH -t 2:00:00 # Run time (hh:mm:ss) #SBATCH --mail-user=mei@usc.edu #SBATCH --mail-type=all #SBATCH -p batch #SBATCH -A geo156 #SBATCH --open-mode=truncate source /ccs/home/mei/scratch/TARGET_UCVM_SFCVM/ucvm_install/conf/ucvm_env.sh srun -N5 -n100 /ccs/home/mei/scratch/sw4-ucvm_cmd/optimize_mp/sw4 sfcvm.txt
making image slice from sfcvm_mesh.sfile
the input file ,sw4_sfcvm.sw4input, needs to update the path links,
# SW4 SCRIPT -- to process sfcvm.mesh.sfile fileio path=output pfs=1 verbose=1 printcycle=10 nwriters=16 # GRID SIZE AND REFINEMENT grid x=280000 y=115000 z=30000 h=100.0 lat=39.111563 lon=-122.569504 az=143.638 proj=tmerc datum=NAD83 lon_p=-123.0 lat_p=35.0 scale=0. 9996 #grid x=20000 y=10000 z=25000 h=200 lat=38.070951 lon=-122.253136 az=143.638 proj=tmerc datum=NAD83 lon_p=-123.0 lat_p=35.0 scale=0.999 6 refinement zmax=3600 #refinement zmax=1500 #refinement zmax=400 supergrid gp=1 time steps=1 globalmaterial vsmin=250 vpmin=500 attenuation phasefreq=1.0 nmech=3 maxfreq=10.0 sfile filename=sfcvm.mesh.sfile directory=/ccs/home/mei/scratch/sw4-ucvm_cmd/ucvm/sfcvm.output #prefilter fc2=2.5 type=lowpass passes=2 order=2 # POINT SOURCE source depth=12300 lat=37.85517 lon=-122.25683 m0=4.563e15 dip=80 strike=145 rake=180 t0=1.0 type=Dirac ## generate image at z depth image mode=s z=0 file=image cycle=0 #image mode=s z=100 file=image cycle=0 #image mode=s z=500 file=image cycle=0 # Stations rec lat=37.59669 lon=-121.88094 depth=0 file=1688_37.59669_-121.88094 variables=velocity nsew=1 usgsformat=1 sacformat=0
and the slurm script to run this
#!/bin/bash #SBATCH -J run_sw4_sfcvm # Job name #SBATCH -o run_sw4_sfcvm.oo # Name of stdout output file #SBATCH -e run_sw4_sfcvm.ee # Name of stderr error file #SBATCH -N 5 # Total # of nodes #SBATCH -n 100 #SBATCH --mem 0 # Set to unlimited memory #SBATCH -t 2:00:00 # Run time (hh:mm:ss) #SBATCH --mail-user=mei@usc.edu #SBATCH --mail-type=all #SBATCH -p batch #SBATCH -A geo156 #SBATCH --open-mode=truncate #SBATCH --export=NONE source /ccs/home/mei/scratch/TARGET_UCVM_SFCVM/ucvm_install/conf/ucvm_env.sh srun -N 5 -n 100 /ccs/home/mei/scratch/sw4-stock/optimize_mp/sw4 ./sw4_sfcvm.sw4input
2 images files are created in 'output' directory:
$ ls output 1688_37.59669_-121.88094.txt image.cycle=00.z=0.s.sw4img image.cycle=0.z=0.s.sw4img
install pySW4,
conda install. pysw4
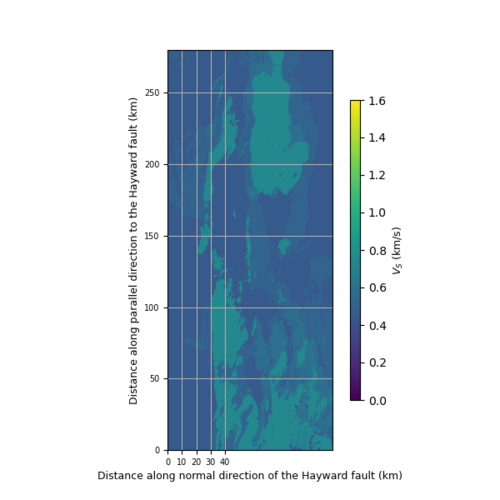
using sw4img_printer.py to generate png files,
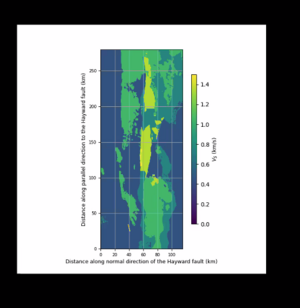
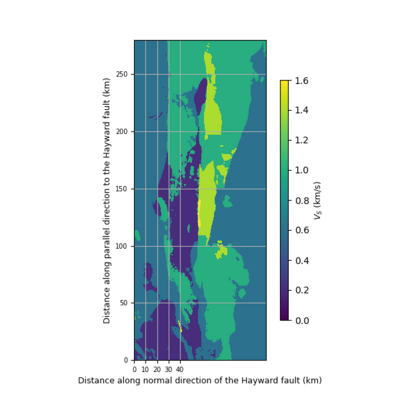
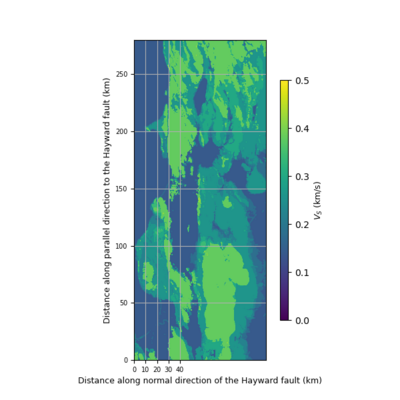
import matplotlib.pyplot as plt
import pySW4 as sw4
import numpy as np
def vm_plain_view(fname, fsw4img, font=9, vmin=0.0, vmax=2.5):
# fsw4img = '../papers/vm_evaluation/vm_img/image.cycle=000.z=0.s.sw4img'
# fsw4img = '../papers/vm_evaluation/vm_img/z0_topo.sw4img'
norm = plt.Normalize(min, vmax)
fig = plt.figure(figsize=(6, 6))
ax = fig.add_axes([0.1, 0.1, 0.8, 0.8])
cbaxes = fig.add_axes([0.7, 0.2, 0.02, 0.6])
fig.colorbar(plt.cm.ScalarMappable(norm=norm, cmap="viridis"), cax=cbaxes).set_label(label='$V_S$ (km/s)', size=font)
cbaxes.set_ylim(vim, vmax)
image = sw4.read_image(fsw4img)
patch = image.patches[0]
x0 = patch.extent[0] * 1e-3
x1 = patch.extent[1] * 1e-3
z0 = patch.extent[2] * 1e-3
z1 = patch.extent[3] * 1e-3
vs = patch.data * 1e-3
vs = np.flip(vs, axis=[1])
x_axis = np.linspace(x0, x1, vs.shape[1])
y_axis = np.linspace(z1, z0, vs.shape[0])
x_mesh, y_mesh = np.meshgrid(x_axis, y_axis)
ax.contourf(x_mesh[::2, ::2], y_mesh[::2, ::2], vs[::2, ::2], norm=norm, cmap='viridis')
ax.set_ylabel('Distance along parallel direction to the Hayward fault (km)', fontsize=font)
ax.grid(which='both')
ax.tick_params(axis='x', labelsize=font - 2)
ax.tick_params(axis='y', labelsize=font - 2)
ax.axis('scaled')
ax.set_xlabel('Distance along normal direction of the Hayward fault (km)', fontsize=font)
box = {'facecolor': 'none',
'edgecolor': 'black',
'boxstyle': 'square'
}
ax.set_xticks([0, 10, 20, 30, 40])
# epstopdf vm_plain_view.eps vm_plain_view.pdf
# plt.savefig('vm_plain_view.pdf', format='pdf')
plt.savefig(fname, format='png')
plt.close()
if __name__ == '__main__':
datapath = 'datapath/image.cycle=00.z=0.s.sw4img'
vm_plain_view('00_view.png', datapath, font=9, vmin=0.0, vmax=0.8)
NOTE : the pySW4 is here, https://github.com/shaharkadmiel/pySW4
Discovery Setup (incomplete)
Currently Loaded Modules:
1) gcc/11.3.0 3) pmix/3.2.3 5) perl/5.36.0 7) openblas/0.3.21 2) openmpi/4.1.4 4) libxml2/2.9.13 6) git/2.36.1
UCVM (withSFCVM branch) at /project/scec_608/ucvm_main_sfcvm
sw4 (ucvm_cmd branch) at /project/scec_608/mei/sw4-ucvm_cmd
using config/make.inc
FC = gfortran CXX = /spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/openmpi-4.1.4-4w23jca/bin/mpiCC proj=yes hdf5=yes SW4ROOT = /project/scec_608/mei/ucvm_main_sfcvm/install/lib/proj SQL3ROOT = /project/scec_608/mei/ucvm_main_sfcvm/install/lib/sqlite HDF5ROOT = /project/scec_608/mei/ucvm_main_sfcvm/install/lib/hdf5 #EXTRA_LINK_FLAGS = -L/usr/lib64 -llapack -lblas -lgfortran EXTRA_LINK_FLAGS = -L/usr/lib64 -lgfortran -L/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/openbla s-0.3.21-afcoykn/lib -lopenblas
sw4 (main branch with v3.0 tag)
mei@discovery1:/project/scec_608/mei/sw4-stock$ git checkout v3.0 Note: switching to 'v3.0'. You are in 'detached HEAD' state. You can look around, make experimental changes and commit them, and you can discard any commits you make in this state without impacting any branches by switching back to a branch. If you want to create a new branch to retain commits you create, you may do so (now or later) by using -c with the switch command. Example: git switch -c <new-branch-name> Or undo this operation with: git switch - Turn off this advice by setting config variable advice.detachedHead to false HEAD is now at 13e6d431 Merge pull request #195 from geodynamics/developer
Study setup
slurm script
#!/bin/bash #SBATCH -J sfcvm_sw4 # Job name #SBATCH -o sfcvm_sw4.oo # Name of stdout output file #SBATCH -e sfcvm_sw4.ee # Name of stderr error file #SBATCH -N 10 # Total # of nodes #SBATCH -n 200 #SBATCH -t 2:00:00 # Run time (hh:mm:ss) #SBATCH --mail-user=mei@usc.edu #SBATCH --mail-type=all #SBATCH -p batch #SBATCH -A geo156 #SBATCH --open-mode=truncate source /ccs/home/mei/scratch/TARGET_UCVM_SFCVM/ucvm_install/conf/ucvm_env.sh srun -N10 -n200 /ccs/home/mei/scratch/sw4-ucvm_cmd/optimize_mp/sw4 sfcvm.txt
26G sfile
sfcvm.txt spec
# SW4 SCRIPT fileio path=sfcvm.output pfs=1 verbose=1 printcycle=10 nwriters=16 # GRID SIZE AND REFINEMENT grid x=280000 y=115000 z=30000 h=200.0 lat=39.111563 lon=-122.569504 az=143.638 proj=tmerc datum=NAD83 lon_p=-123.0 lat_p=35.0 scale=0.9996 refinement zmax=3600 refinement zmax=1500 #refinement zmax=400 supergrid gp=1 time t=0.1 utcstart=01/01/2021:00:00:00.0 #globalmaterial vsmin=250 vpmin=500 attenuation phasefreq=1.0 nmech=3 maxfreq=10.0 ##sfile filename=sfcvm.mesh.sfile directory=/lustre/orion/geo156/proj-shared/pinillar/sw4_test ucvm filename=sfcvm.h200 #prefilter fc2=2.5 type=lowpass passes=2 order=2 # POINT SOURCE source depth=12300 lat=37.85517 lon=-122.25683 m0=4.563e15 dip=80 strike=145 rake=180 t0=1.0 type=Dirac image mode=s z=0 file=image cycle=0 # Stations rec lat=37.59669 lon=-121.88094 depth=0 file=1688_37.59669_-121.88094 variables=velocity nsew=1 usgsformat=1 sacfor mat=0 #rec x=10000 y=10000 z=0 file=rec_11500 variables=velocity nsew=1 usgsformat=1 sacformat=0 sfileoutput file=sfcvm.mesh
sw4 result log
11G sfile
sfcvm.txt spec
# SW4 SCRIPT fileio path=sfcvm.output pfs=1 verbose=1 printcycle=10 nwriters=16 # GRID SIZE AND REFINEMENT grid x=280000 y=115000 z=30000 h=200.0 lat=39.111563 lon=-122.569504 az=143.638 proj=tmerc datum=NAD83 lon_p=-123.0 lat_p=35.0 scale=0.9996 refinement zmax=3600 refinement zmax=1500 #refinement zmax=400 supergrid gp=1 time t=0.1 utcstart=01/01/2021:00:00:00.0 #globalmaterial vsmin=250 vpmin=500 attenuation phasefreq=1.0 nmech=3 maxfreq=10.0 ##sfile filename=sfcvm.mesh.sfile directory=/lustre/orion/geo156/proj-shared/pinillar/sw4_test ucvm filename=sfcvm.h200 #prefilter fc2=2.5 type=lowpass passes=2 order=2 # POINT SOURCE source depth=12300 lat=37.85517 lon=-122.25683 m0=4.563e15 dip=80 strike=145 rake=180 t0=1.0 type=Dirac image mode=s z=0 file=image cycle=0 # Stations rec lat=37.59669 lon=-121.88094 depth=0 file=1688_37.59669_-121.88094 variables=velocity nsew=1 usgsformat=1 sacfor mat=0 #rec x=10000 y=10000 z=0 file=rec_11500 variables=velocity nsew=1 usgsformat=1 sacformat=0 sfileoutput file=sfcvm.mesh
sw4 result log,
----------------------------------------------------------------
sw4 version 3.0
This program comes with ABSOLUTELY NO WARRANTY; released under GPL.
This is free software, and you are welcome to redistribute
it under certain conditions, see LICENSE.txt for more details
----------------------------------------------------------------
Compiled on: Fri 26 Jul 2024 06:46:45 PM EDT
By user: mei
Machine: login08
Compiler: /opt/cray/pe/mpich/8.1.27/ofi/gnu/9.1/bin/mpicxx
3rd party include dir: /ccs/home/mei/scratch/TARGET_UCVM_SFCVM/ucvm_install/lib/proj/include, and library dir:
/ccs/home/mei/scratch/TARGET_UCVM_SFCVM/ucvm_install/lib/proj/lib
----------------------------------------------------------------
Input file: sfcvm.txt
Default Supergrid thickness has been tuned; # grid points = 1 grid sizes
Default Supergrid damping coefficient has been tuned; damping coefficient = 2.00000000e-02
* Processing the grid command...
* Setting nx to 1401 to be consistent with h=2.00000000e+02
* Setting ny to 576 to be consistent with h=2.00000000e+02
* Setting nz to 151 to be consistent with h=2.00000000e+02
cleanupRefinementLevels: topo_zmax = 0.00000000e+00
Cartesian refinement levels (z=):
3.60000000e+03
1.50000000e+03
0.00000000e+00
Grid distributed on 200 processors
Finest grid size 5607 x 2307
Processor array 20 x 10
Number of curvilinear grids = 0
Number of Cartesian grids = 3
Total number of grids = 3
Extent of the computational domain xmax=2.80000000e+05 ymax=1.15000000e+05 zmax=3.00000000e+04
Cartesian refinement levels after correction:
Grid=0 z-min=3.60000000e+03
Grid=1 z-min=1.50000000e+03
Grid=2 z-min=0.00000000e+00
Corrected global_zmax = 3.00000000e+04
Rank=0, Grid #2 (Cartesian), iInterior=[1,281], jInterior=[1,231], kInterior=[1,31]
Rank=0, Grid #1 (Cartesian), iInterior=[1,141], jInterior=[1,116], kInterior=[1,22]
Rank=0, Grid #0 (Cartesian), iInterior=[1,71], jInterior=[1,58], kInterior=[1,133]
*** No topography command found in input file. Using z=0 as free surface boundary ***
Global grid sizes (without ghost points)
Grid h Nx Ny Nz Points Type
0 200 1401 576 133 107327808 Cartesian
1 100 2801 1151 22 70926922 Cartesian
2 50 5601 2301 31 399524931 Cartesian
Total number of grid points (without ghost points): 5.7778e+08
*** Using UCVM sfcvm.h200 in directory ./
MaterialUCVM::read_UCVM, time to read material file: 3.01000000e-07 seconds.
Cartesian coordinates of source at (lon, lat)=(-1.222568e+02, 3.785517e+01) is (x,y)=(128873, 59901.3)
Read station input, took 2.35780000e-04seconds.
Execution time, reading input file 2.60684555e-01 seconds
Assuming a PARALLEL file system
Writing images from (up to) 16 procs
Detected at least one boundary with supergrid conditions
----------------------------------------------------
Making Directory: sfcvm.output/
... Done!
----------------------------------------------------
Geographic and Cartesian coordinates of the corners of the computational grid:
0: Lon= -1.225695e+02, Lat=3.911156e+01, x=5.624866e-10, y=4.141253e-10
1: Lon= -1.207151e+02, Lat=3.705896e+01, x=2.800000e+05, y=1.164153e-09
2: Lon= -1.217658e+02, Lat=3.646019e+01, x=2.800000e+05, y=1.150000e+05
3: Lon= -1.236348e+02, Lat=3.849645e+01, x=-8.294592e-10, y=1.150000e+05
top_right = [39.111563, -122.569504]
bot_right = [37.058959, -120.715088]
bot_left = [36.460193, -121.765788]
top_left = [38.496453, -123.634753]
Considering all material blocks
Query last batch 28
Read 572402 points
Rank 0 grid 0: rho min = -1.00, max = 3300.00
Rank 0 grid 0: cp min = -1.00, max = 7781.14
Rank 0 grid 0: cs min = -1.00, max = 4416.69
Query last batch 22
Read 457968 points
Rank 0 grid 1: rho min = -1.00, max = 2663.03
Rank 0 grid 1: cp min = -1.00, max = 5486.68
Rank 0 grid 1: cs min = -1.00, max = 3294.37
Query last batch 120
Read 2401707 points
Rank 0 grid 2: rho min = -1.00, max = 2656.13
Rank 0 grid 2: cp min = -1.00, max = 4834.72
Rank 0 grid 2: cs min = -1.00, max = 2904.44
UCVM command: outside = 0, material = 476852337
----------- Material properties ranges ---------------
1.60438900e+03 kg/m^3 <= Density <= 3.30000000e+03 kg/m^3
7.30335000e+02 m/s <= Vp <= 7.78155700e+03 m/s
8.02080000e+01 m/s <= Vs <= 4.41693400e+03 m/s
1.46159672e+00 <= Vp/Vs <= 1.49239438e+01
1.03217139e+07 Pa <= mu <= 6.43807097e+10 Pa
5.47929452e+07 Pa <= lambda <= 7.10622575e+10 Pa
Using attenuation
4.01040000e+00 <= Qs <= 2.20846700e+02
8.02080000e+00 <= Qp <= 4.41693400e+02
------------------------------------------------------
***** PPW = minVs/h/maxFrequency ********
g=0, h=2.000000e+02, minVs/h=9.02643 (Cartesian)
g=1, h=1.000000e+02, minVs/h=8.72043 (Cartesian)
g=2, h=5.000000e+01, minVs/h=1.60416 (Cartesian)
*** Attenuation parameters calculated for 3 mechanisms,
max freq=1.000000e+01 [Hz], min_freq=1.000000e-01 [Hz], velo_freq=1.000000e+00 [Hz]
omega[0]=6.283185e-01 omega[1]=6.283185e+00 omega[2]=6.283185e+01
omc[0]=6.283185e-01 omc[1]=1.986918e+00 omc[2]=6.283185e+00 omc[3]=1.986918e+01 omc[4]=6.283185e+01
Assigned material properties
*** computing the time step ***
TIME accuracy order=4 CFL=1.30000000e+00 prel. time step=8.72078955e-03
time step = 9.09090909e-03
*** Initialized Images
Min source z-level: 1.230000e+04, max source z-level: 1.230000e+04
Execution time, start up phase 52 minutes 3.78607278e+01 seconds
Running sw4 on 200 processors...
Writing output to directory: sfcvm.output/
*** Starting solve ***
============================================================
Running program on 200 MPI tasks using the following data:
Start Time = 0 Goal Time = 0.1
Number of time steps = 11 dt: 0.00909091
============================================================
Global boundary conditions
x=0 supergrid
x=xMax supergrid
y=0 supergrid
y=yMax supergrid
z=topo free surface
z=zMax supergrid
Query last batch 28
..
22G sfile
sfcvm.txt spec
# SW4 SCRIPT fileio path=sfcvm.output pfs=1 verbose=1 printcycle=10 nwriters=16 # GRID SIZE AND REFINEMENT grid x=280000 y=115000 z=30000 h=400.0 lat=39.111563 lon=-122.569504 az=143.638 proj=tmerc datum=NAD83 lon_p=-123.0 lat_p=35.0 scale=0.9996 refinement zmax=10000 refinement zmax=3600 refinement zmax=1300 refinement zmax=350 supergrid gp=1 time t=0.1 utcstart=01/01/2021:00:00:00.0 #globalmaterial vsmin=250 vpmin=500 attenuation phasefreq=1.0 nmech=3 maxfreq=10.0 ##sfile filename=sfcvm.mesh.sfile directory=/lustre/orion/geo156/proj-shared/pinillar/sw4_test ucvm filename=sfcvm.h200 #prefilter fc2=2.5 type=lowpass passes=2 order=2 # POINT SOURCE source depth=12300 lat=37.85517 lon=-122.25683 m0=4.563e15 dip=80 strike=145 rake=180 t0=1.0 type=Dirac image mode=s z=0 file=image cycle=0 # Stations rec lat=37.59669 lon=-121.88094 depth=0 file=1688_37.59669_-121.88094 variables=velocity nsew=1 usgsformat=1 sacfor mat=0 #rec x=10000 y=10000 z=0 file=rec_11500 variables=velocity nsew=1 usgsformat=1 sacformat=0 sfileoutput file=sfcvm.mesh
sw4 result log
sw4 plots (22G)
Validate with depth profiles
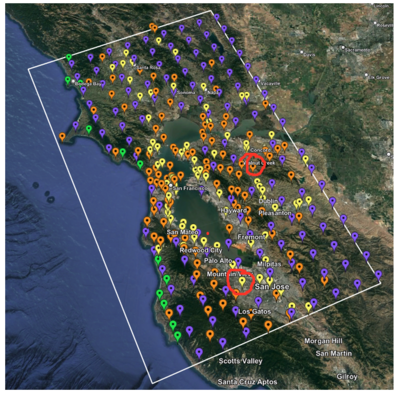
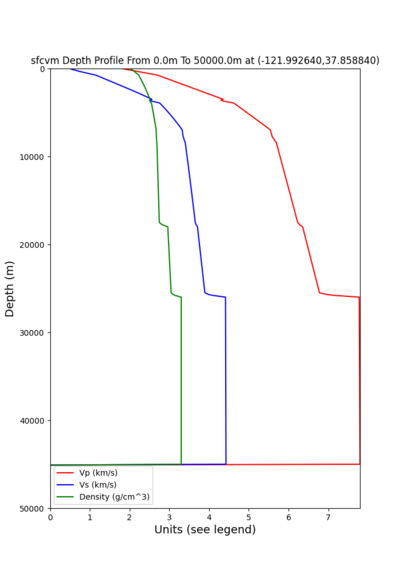
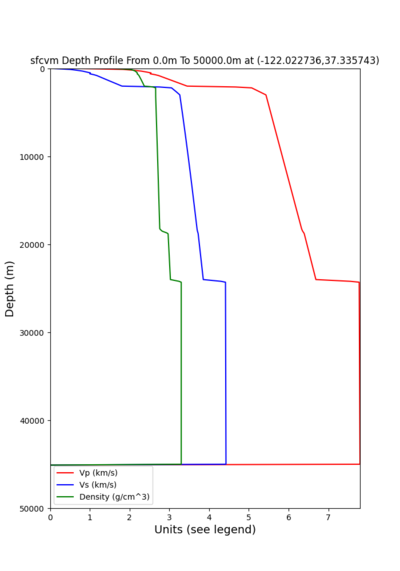
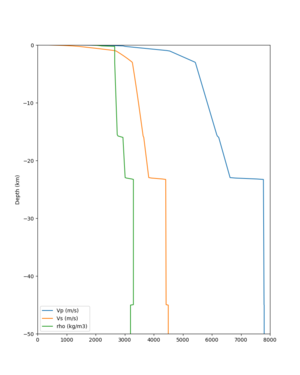
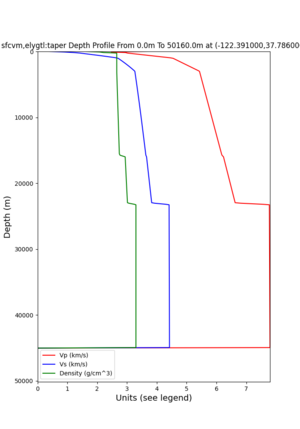
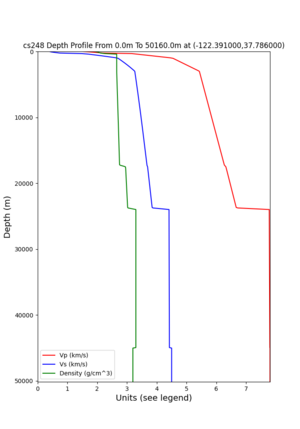
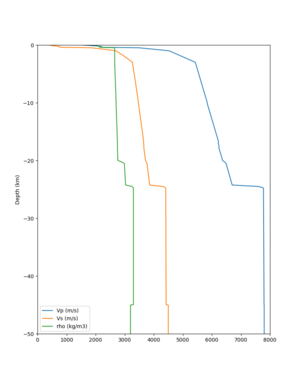
2 cybershake sites:
RNHL : 3,RNHL,,37.858840,-121.992640 CFCS : 1,CFCS,,37.335743,-122.022736
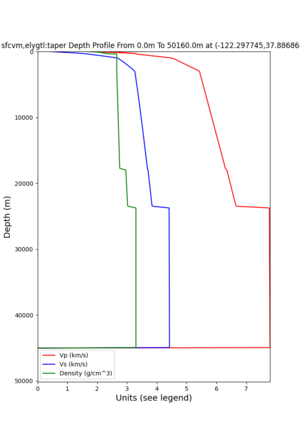
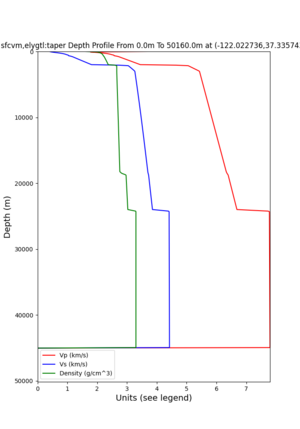
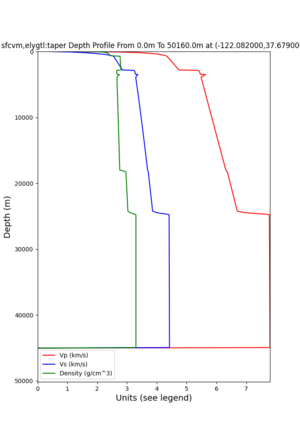
plot_depth_profile.py -s 37.858840,-121.992640 -b 0 -e 50000 -d vs,vp,density -v 100 -c sfcvm -o depth_RNHL .png -i /var/www/html/UCVM_web_sfcvm/web/model/UCVM_TARGET plot_depth_profile.py -s 37.335743,-122.022736 -b 0 -e 50000 -d vs,vp,density -v 100 -c sfcvm -o depth_CFCS .png -i /var/www/html/UCVM_web_sfcvm/web/model/UCVM_TARGET
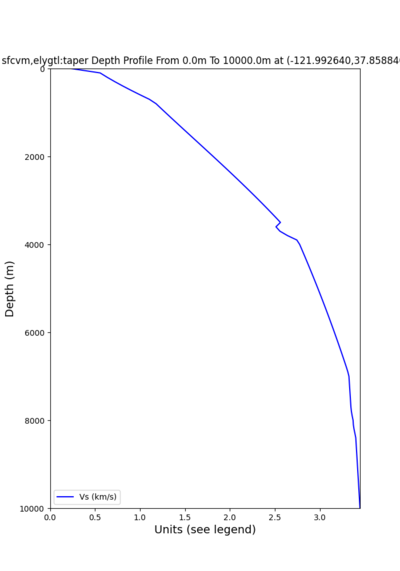
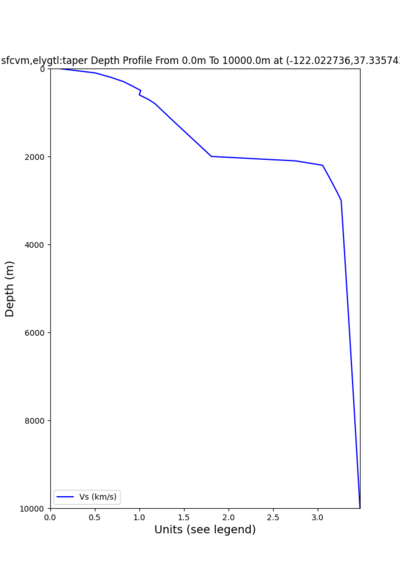
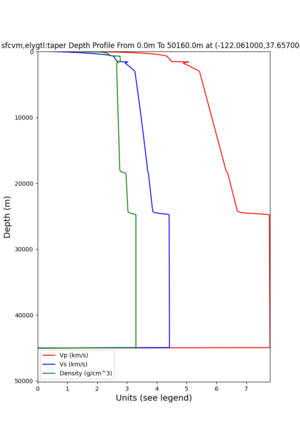
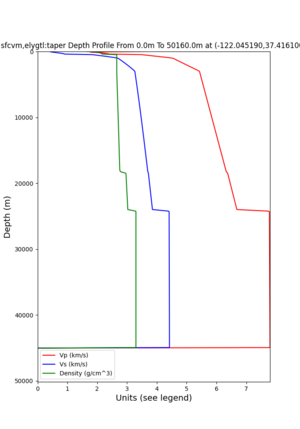
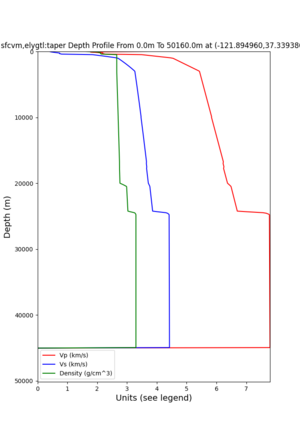
plot_depth_profile.py -s 37.858840,-121.992640 -b 0 -e 10000 -d vs -v 100 -c sfcvm,elygtl:taper -o depth_RN HL_1.png -i /var/www/html/UCVM_web_sfcvm/web/model/UCVM_TARGET -L 100,500,500 plot_depth_profile.py -s 37.335743,-122.022736 -b 0 -e 10000 -d vs -v 100 -c sfcvm,elygtl:taper -o depth_CF CS_1.png -i /var/www/html/UCVM_web_sfcvm/web/model/UCVM_TARGET -L 100,500,500
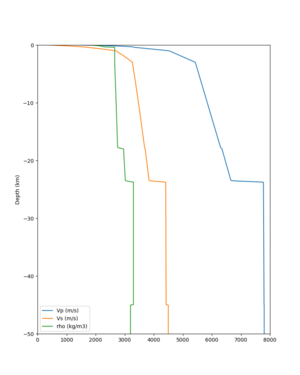
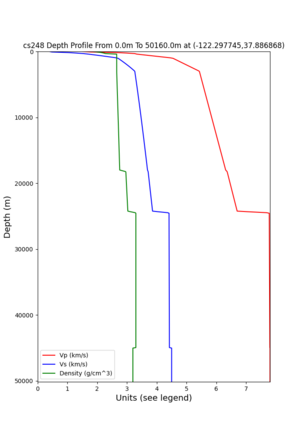
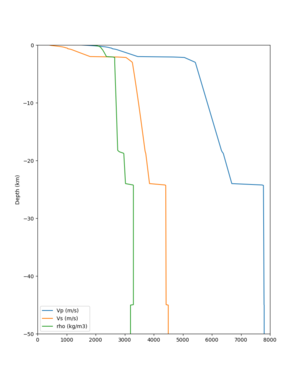
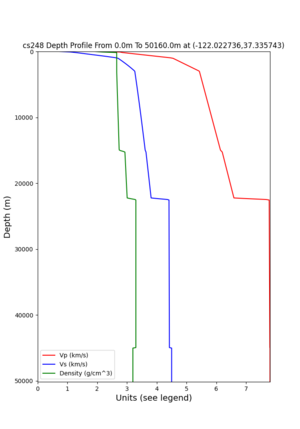
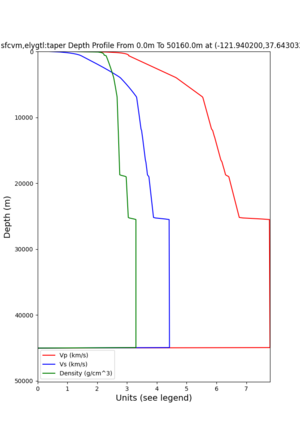
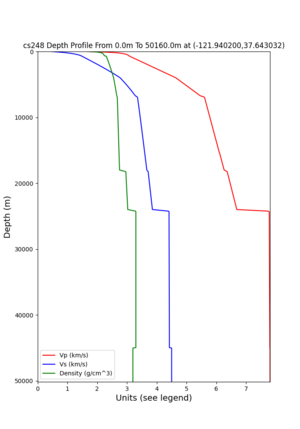
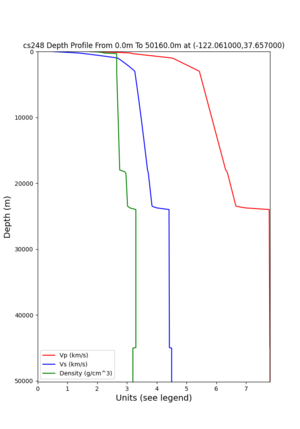
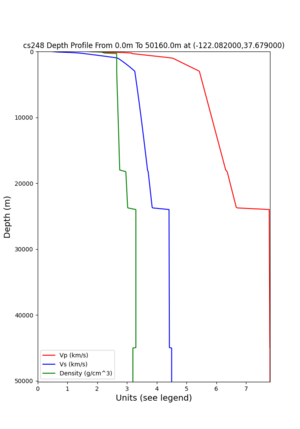
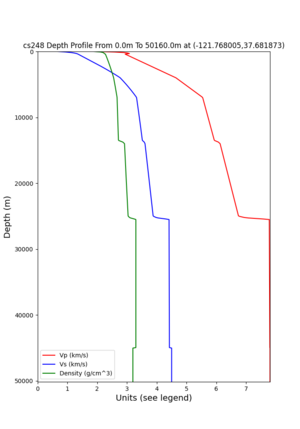
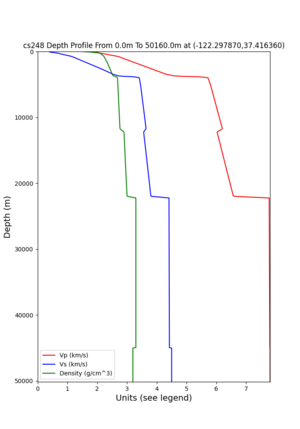
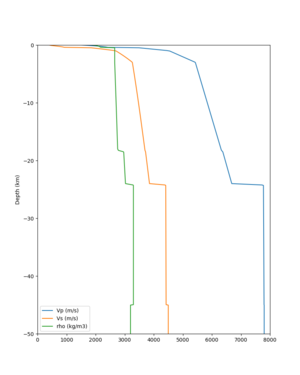
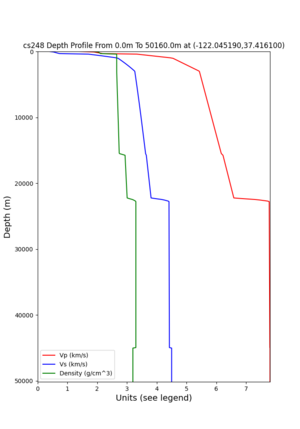
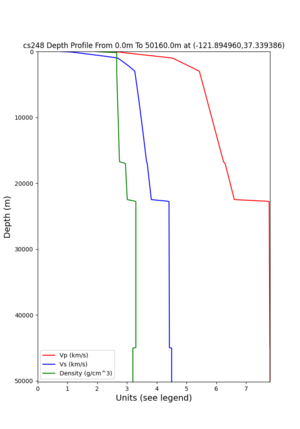
cs248 plots through UCVM
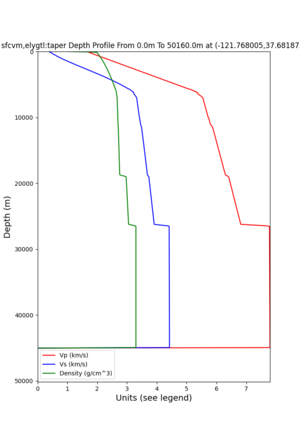
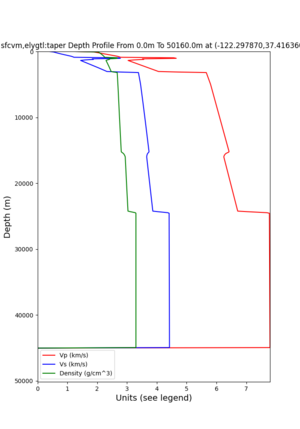
Site,Lat,Lon, ALBY,37.886868,-122.297745, CFCS,37.335743,-122.022736, CSU1,37.643032,-121.9402, CSUEB,37.657,-122.061, HAYW,37.679,-122.082, LVMR,37.681873,-121.768005, s3171,37.41636,-122.29787, s3240,37.4161,-122.04519, SFRH,37.786,-122.391, SJO,37.339386,-121.89496,
ALBY
CFCS
CSU1
CSUEB
HAYW
LVMR
s3171
s3240
SFRH
SJO
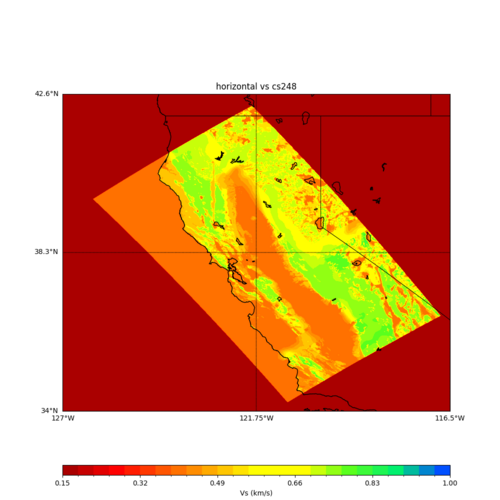
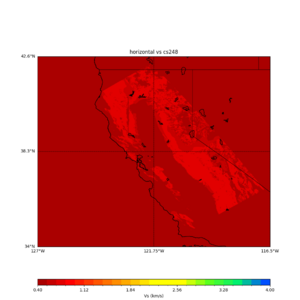
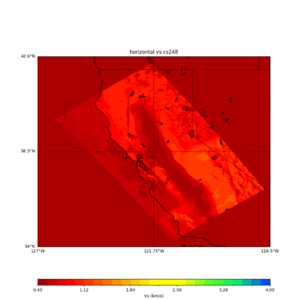
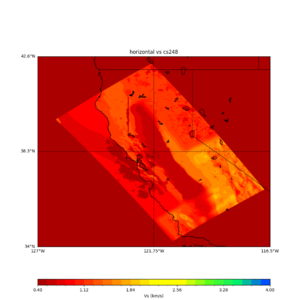
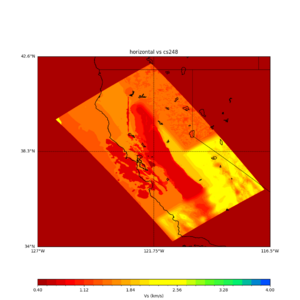
sw4 slice of cs248
ucvm vs slices of cs248
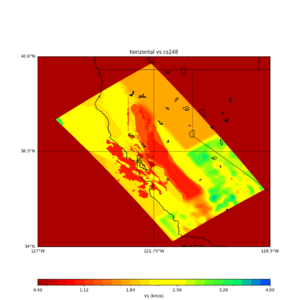
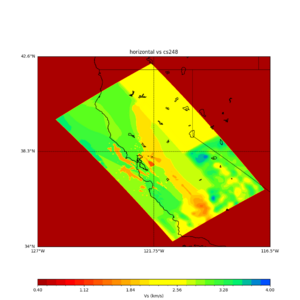
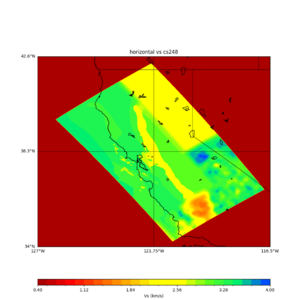
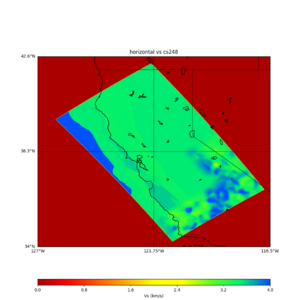
plot_horizontal_slice.py -s 0.01 -c cs248 -a d -o cs248_vs.png -i $UCVM_INSTALL_PATH -t "horizontal vs cs248" -d vs -e 0 -b 34,-127 -u 42.6,-116.5 -A 0.15,1.0
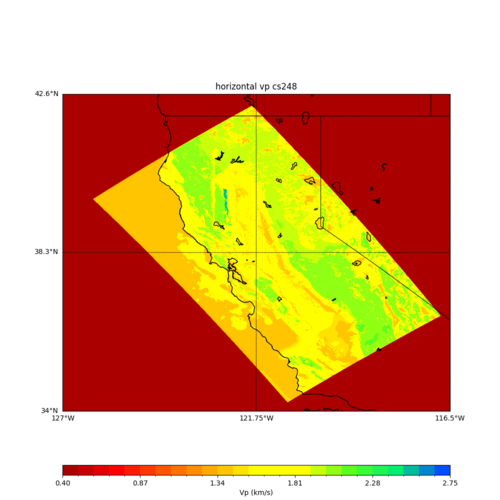
ucvm vp slice of cs248
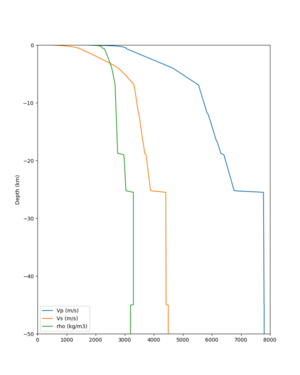
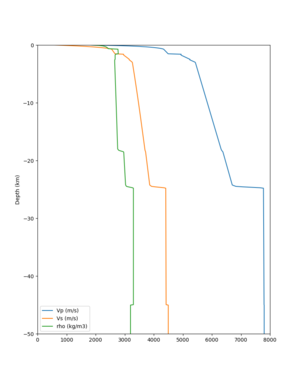
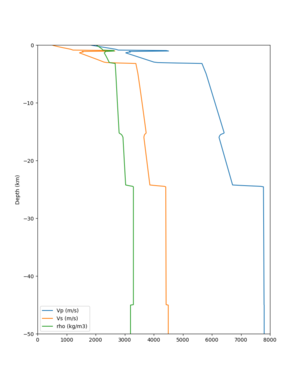
Scott's study of s3171
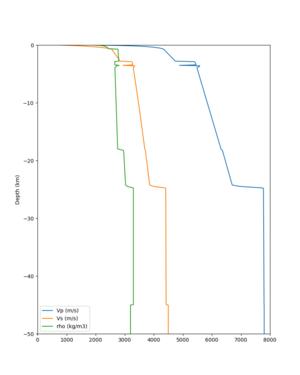
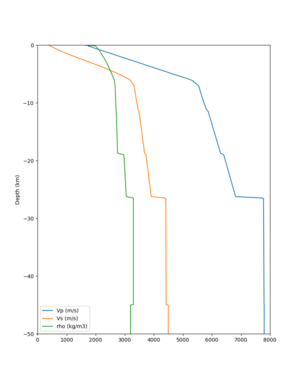
I wanted to provide an update on my investigation of the velocity model differences Mei found. I dug into s3171. These are the values from the CyberShake mesh: Depth Vs -0.8 1178.842041 -0.88 1220.528809 -0.96 2603.321777 -1.04 2661.860596 -1.12 1535.679932 -1.2 1575.72937 So something weird is happening around 1 km depth. I queried UCVM using ucvm_query and got similar results (Vs values in bold): [callag@login12.frontier bin]$ ./ucvm_query -f ../conf/ucvm.conf -m sfcvm Using Geo Depth coordinates as default mode. -122.29787 37.41636 800.0 -122.29787 37.41636 850.0 -122.29787 37.41636 900.0 -122.29787 37.41636 950.0 -122.29787 37.41636 1000.0 -122.29787 37.41636 1050.0 -122.29787 37.41636 1100.0 -122.29787 37.41636 1150.0 -122.2979 37.4164 800.000 561.816 610.603 sfcvm 2735.530 1181.555 2237.740 none 0.000 0.000 0.000 crust 2735.530 1181.555 2237.740 -122.2979 37.4164 850.000 561.816 610.603 sfcvm 2765.738 1207.573 2243.892 none 0.000 0.000 0.000 crust 2765.738 1207.573 2243.892 -122.2979 37.4164 900.000 561.816 610.603 sfcvm 2795.947 1233.704 2249.995 none 0.000 0.000 0.000 crust 2795.947 1233.704 2249.995 -122.2979 37.4164 950.000 561.816 610.603 sfcvm 4550.641 2707.392 2667.763 none 0.000 0.000 0.000 crust 4550.641 2707.392 2667.763 -122.2979 37.4164 1000.000 561.816 610.603 sfcvm 4633.183 2766.236 2667.770 none 0.000 0.000 0.000 crust 4633.183 2766.236 2667.770 -122.2979 37.4164 1050.000 561.816 610.603 sfcvm 4655.962 2782.238 2667.778 none 0.000 0.000 0.000 crust 4655.962 2782.238 2667.778 -122.2979 37.4164 1100.000 561.816 610.603 sfcvm 3508.835 1828.182 2406.959 none 0.000 0.000 0.000 crust 3508.835 1828.182 2406.959 -122.2979 37.4164 1150.000 561.816 610.603 sfcvm 3536.539 1850.997 2410.829 none 0.000 0.000 0.000 crust 3536.539 1850.997 2410.829 Next I used geomodelgrids_query: [callag@login12.frontier bin]$ cat s3171_pt.in 37.4164 -122.2979 -800.0 37.4164 -122.2979 -850.0 37.4164 -122.2979 -900.0 37.4164 -122.2979 -950.0 37.4164 -122.2979 -1000.0 37.4164 -122.2979 -1050.0 37.4164 -122.2979 -1100.0 37.4164 -122.2979 -1150.0 [callag@login12.frontier ./geomodelgrids_query --models=../../../data/sfcvm/USGS_SFCVM_v21-1_detailed.h5 --points=s3171_pt.in --output=s3171_pt.out --values=vs cat s3171_pt.out # /lustre/orion/geo156/proj-shared/CyberShake/software/UCVM/ucvm/work/model/sfcvm/dependencies/geomodelgrids-build/bin/.libs/geomodelgrids_query --models=../../../data/sfcvm/USGS_SFCVM_v21-1_detailed.h5 --points=s3171_pt.in --output=s3171_pt.out --values=vs # x0 x1 x2 vs 3.741640e+01 -1.222979e+02 -8.000000e+02 1.426508e+03 3.741640e+01 -1.222979e+02 -8.500000e+02 1.453045e+03 3.741640e+01 -1.222979e+02 -9.000000e+02 1.479618e+03 3.741640e+01 -1.222979e+02 -9.500000e+02 1.506220e+03 3.741640e+01 -1.222979e+02 -1.000000e+03 1.532844e+03 3.741640e+01 -1.222979e+02 -1.050000e+03 1.559482e+03 3.741640e+01 -1.222979e+02 -1.100000e+03 1.586127e+03 3.741640e+01 -1.222979e+02 -1.150000e+03 1.612773e+03 You'll notice these values are quite different. However, if I repeat the query with a squashing depth of 45 km, which I think is what UCVM is using: [callag@login12.frontier bin]$ ./geomodelgrids_query --models=../../../data/sfcvm/USGS_SFCVM_v21-1_detailed.h5 --points=s3171_pt.in --output=s3171_pt.out --squash-min-elev=-4.5e+4 --values=vs [callag@login12.frontier bin]$ cat s3171_pt.out # /lustre/orion/geo156/proj-shared/CyberShake/software/UCVM/ucvm/work/model/sfcvm/dependencies/geomodelgrids-build/bin/.libs/geomodelgrids_query --models=../../../data/sfcvm/USGS_SFCVM_v21-1_detailed.h5 --points=s3171_pt.in --output=s3171_pt.out --squash-min-elev=-4.5e+4 --values=vs # x0 x1 x2 vs 3.741640e+01 -1.222979e+02 -8.000000e+02 1.181600e+03 3.741640e+01 -1.222979e+02 -8.500000e+02 1.207619e+03 3.741640e+01 -1.222979e+02 -9.000000e+02 1.233750e+03 3.741640e+01 -1.222979e+02 -9.500000e+02 2.707678e+03 3.741640e+01 -1.222979e+02 -1.000000e+03 2.768086e+03 3.741640e+01 -1.222979e+02 -1.050000e+03 2.784075e+03 3.741640e+01 -1.222979e+02 -1.100000e+03 1.868311e+03 3.741640e+01 -1.222979e+02 -1.150000e+03 1.890836e+03 then we get values quite similar to what UCVM is seeing, with this big jump around 1 km depth. As a result, I think this is a squashing artifact, which we should report but not necessarily worry about fixing for this study. Please let me know if you feel differently.
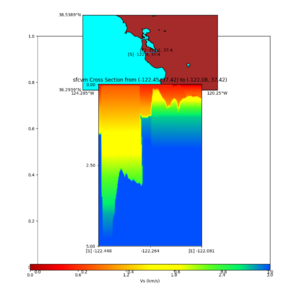
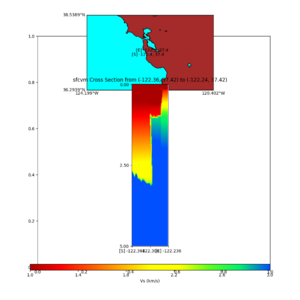
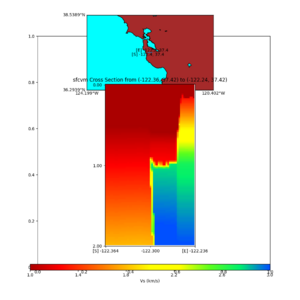
additional plots at s3171
plot_cross_section.py -s 0 -h 204 -d vs -c sfcvm -a sd -o sfcvm_vs_s3171_5k_l_c.png -i $UCVM_INSTALL_PATH -v 20 -b 37.4164,-122.448 -u 37.4164,-122.081 -e 5000 plot_cross_section.py -s 0 -h 204 -d vs -c cs248 -a sd -o cs248_vs_s3171_5k_l_c.png -i $UCVM_INSTALL_PATH -v 20 -b 37.4164,-122.448 -u 37.4164,-122.081 -e 5000 plot_cross_section.py -s 0 -h 204 -d vs -c sfcvm -a sd -o sfcvm_vs_s3171_5k_c.png -i $UCVM_INSTALL_PATH -v 2 0 -b 37.4164,-122.3637 -u 37.4164,-122.2359 -e 5000 plot_cross_section.py -s 0 -h 204 -d vs -c cs248 -a sd -o cs248_vs_s3171_5k_c.png -i $UCVM_INSTALL_PATH -v 2 0 -b 37.4164,-122.3637 -u 37.4164,-122.2359 -e 5000 plot_cross_section.py -s 0 -h 204 -d vs -c sfcvm -a sd -o sfcvm_vs_s3171_2k_c.png -i $UCVM_INSTALL_PATH -v 2 0 -b 37.4164,-122.3637 -u 37.4164,-122.2359 -e 2000 plot_cross_section.py -s 0 -h 204 -d vs -c cs248 -a sd -o cs248_vs_s3171_2k_c.png -i $UCVM_INSTALL_PATH -v 2 0 -b 37.4164,-122.3637 -u 37.4164,-122.2359 -e 2000