UCVM svm1d
From SCECpedia
GTL
Adding a new SVM gtl for UCVMC
- svmgtl
depth profiles
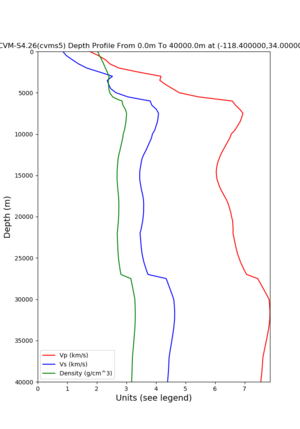
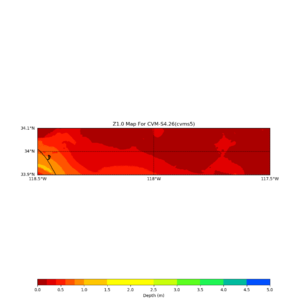
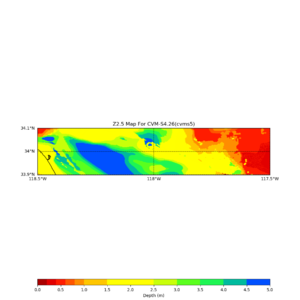
Target point: -118.4,34
echo "-118.4 34.0 " | basin_query -m cvms5 -f ../conf/ucvm.conf
returns the Z1.0 at 580.0
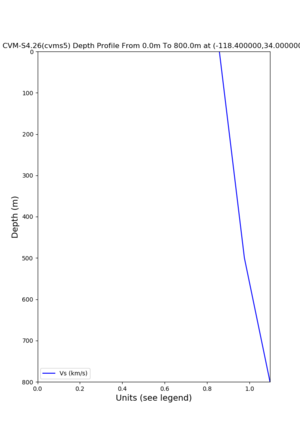
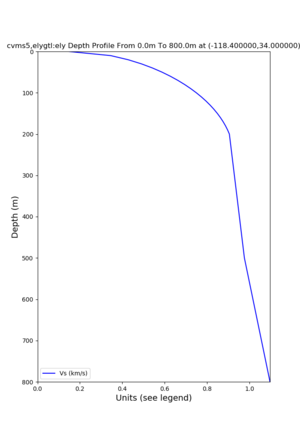
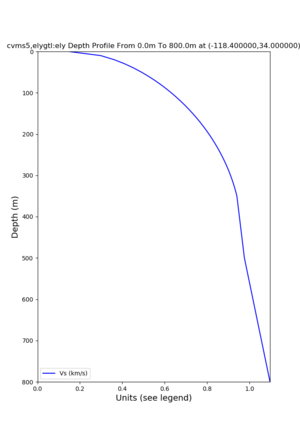
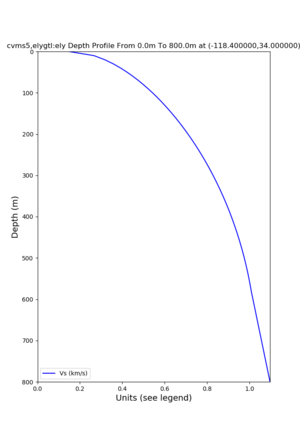
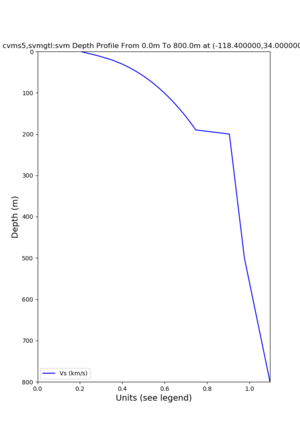
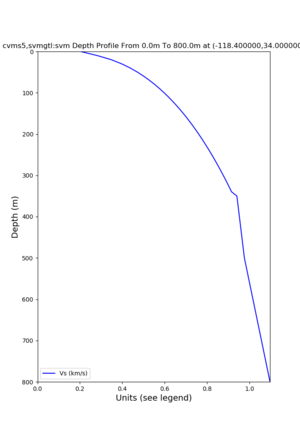
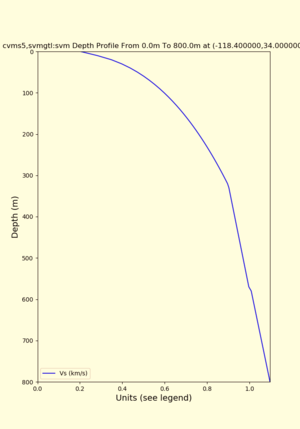
Profile plots,
commands used :
./plot_depth_profile.py -s 34,-118.4 -b 0 -e 40000 -d vs,vp,density -v 100 -c cvms5 -o cvms5_depth_nogtl_bkg.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 800 -d vs -v 10 -c cvms5 -o cvms5_depth_nogtl_base.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 800 -d vs -v 10 -c cvms5,elygtl:ely -z 0,200 -o cvms5_depth_elygtl_200.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 800 -d vs -v 10 -c cvms5,elygtl:ely -z 0,350 -o cvms5_depth_elygtl_350.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 800 -d vs -v 10 -c cvms5,elygtl:ely -Z 1000 -o cvms5_depth_elygtl_Z1.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 800 -d vs -v 10 -c cvms5,svmgtl:svm -z 0,200 -o cvms5_depth_svmgtl_200.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 800 -d vs -v 10 -c cvms5,svmgtl:svm -z 0,350 -o cvms5_depth_svmgtl_350.png ./plot_depth_profile.py -s 34,-118.4 -b 0 -e 800 -d vs -v 10 -c cvms5,svmgtl:svm -Z 1000 -o cvms5_depth_svmgtl_Z1.png
Somewhere by the water
Near westside with cvms5, internal GTL disabled,
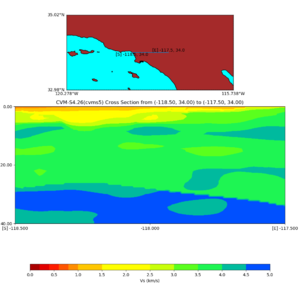
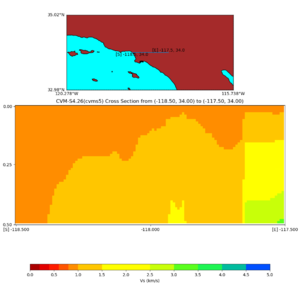
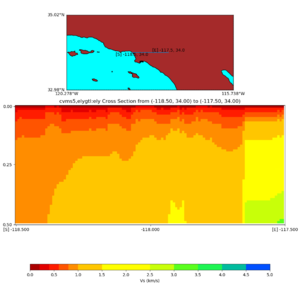
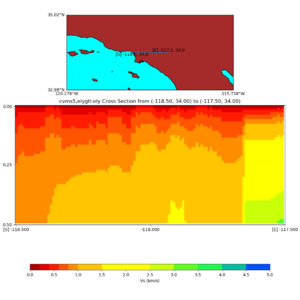
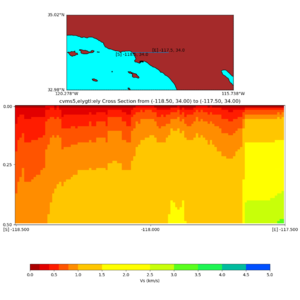
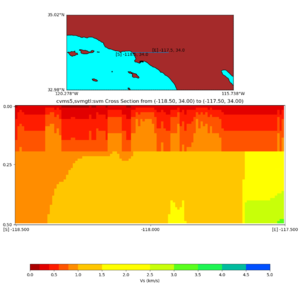
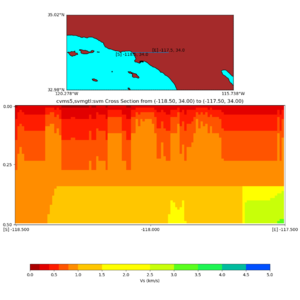
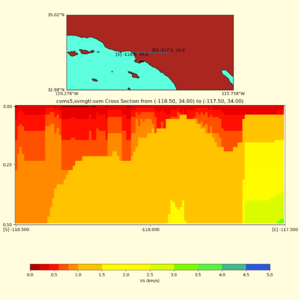
Depth cross plot,
sample commands used :
./plot_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 700 -v 10 -d vs -c cvms5 -a d -s 0 -e 800 -o cvms5_cross_base.png
and,
./plot_cross_section.py -b 34,-118.5 -u 34,-117.5 -h 500 -v 10 -d vs -c cvms5,svmgtl:svm -a d -Z 1000 -s 0 -e 800 -o cvms5_cross_svmgtl_Z1.png
Additional Info
SVM interpolation function extracted by Elnaz from Jian's class_svm.py from https://github.com/jsh9/PySeismoSoil
// zmax would be the Z1.0 if there is one and vs30
// in data->gtl.vs
double calc_z1_from_Vs30(double vs30) {
double z1 = 140.511*exp(-0.00303*vs30); // [m]
return z1;
}
double calc_rho (double vs, double z) {
if (z == 0.0) {
z = 0.0001;
}
double lb = 1.65; //lower bound [g/cm^3]
double rho = 1000.0 * max(lb, 1.0 + 1.0/ (0.614 + 58.7 * (log(z) + 1.095) / vs)); //[kg/m^3]
return rho;
}
/* SVM interpolation method */
int ucvm_interp_svm(double zmin, double zmax, ucvm_ctype_t cmode, ucvm_point_t *pnt, ucvm_data_t *data) {
// curve fitting parameters for SVM model
double p1 = -2.1688E-04;
double p2 = 0.5182 ;
double p3 = 69.452 ;
double r1 = -59.67 ;
double r2 = -0.2722 ;
double r3 = 11.132 ;
double s1 = 4.110 ;
double s2 = -1.0521E-04;
double s3 = -10.827 ;
double s4 = -7.6187E-03;
double zstar = 2.5 ; // [m]
double z1 = zmax; // z at which vs = 1000
double vz1;
double z = data->depth; // interpolation depth
double vs, k, n, vs0; // vs profiling parameters
double vscap = 1000.0;
double eta = 0.9 ;
double zeta, veta ;
double vs30 = data->gtl.vs;
double nu = 0.3; // poisson ratio
double vp_vs = sqrt(2.0*(1.0-nu)/(1.0-2.0*nu));
//~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
switch (cmode) {
case UCVM_COORD_GEO_DEPTH:
case UCVM_COORD_GEO_ELEV:
break;
default:
fprintf(stderr, "Unsupported coord type\n");
return(UCVM_CODE_ERROR);
break;
}
if (z < 0.0) {
return(UCVM_CODE_NODATA);
}
// if no z1 data, compute empirically
if (z1 == 0.0 || z1 == -1.0) {
z1 = calc_z1_from_Vs30(vs30);
}
// query in crustal properties
if (z >= z1) {
data->cmb.vp = data->crust.vp;
data->cmb.vs = data->crust.vs;
data->cmb.rho = data->crust.rho;
data->cmb.source = UCVM_SOURCE_CRUST;
return(UCVM_CODE_SUCCESS);
}
// z is between 0 and z1: query in SVM model
vs0 = p1*pow(vs30,2.0) + p2*vs30 + p3;
if (z1<=zstar) {
vs = min(vs0,vscap);
}
else { // z1 > zstar
k = exp(r1*pow(vs30,2.0) + r3); // <<=== r2 ???
n = max(1.0, s1*exp(s2*vs30) + s3*exp(s4*vs30));
vz1 = vs0*pow(1.0+k*(z1-zstar),1.0/n); // vs @ z1
if (vz1 <= vscap) { // no need to cap the model
if (z<=zstar) {
vs = vs0;
}
else {
vs = vs0*pow(1.0+k*(z-zstar),1.0/n);
}
}
else { // vz1 > vscap -> need to cap the model with linear interpolation from zeta to z1
veta = eta*vscap;
zeta = (1.0/k)*(pow(veta/vs0,n)-1.0)+zstar; // depth at which vs = eta*vscap
if (z <= zeta) {
if (z<=zstar) {
vs = vs0;
}
else {
vs = vs0*pow(1.0+k*(z-zstar),1.0/n);
}
}
else { // z>zeta -> linear interpolation
vs = veta + ((vcap-veta)/(z1-zeta))*(z-zeta);
}
}
}
data->cmb.vs = vs;
data->cmb.vp = vp_vs * vs;
data->cmb.rho = calc_rho(vs,z);
data->cmb.source = data->gtl.source;
return(UCVM_CODE_SUCCESS);
}