UCVM cvmsi tapering for CyberShake Study 22.12
From SCECpedia
Revision as of 23:06, 20 March 2023 by Mei (talk | contribs) (→Create difference map between 2 binary files)
Contents
UCVM tapering
interp Zrange is set to [0,700] initially
related ucvm_query flags that affects tapering:
-L vs,vp,density (optional)
example, -L 200,-1,-1
(only floor the vs to 200)
-m cvmsi,elygtl:taper
(adding tapering interp to cvmsi)
-z zmin,zmax (optional)
(zrange values, original default: zmin = 0, zmax = 350)
UCVM's implementation of ucvm-tapering is ..
zmax fixed at 700
outside of zrange
below zmin, process GTL (vs30 to vs)
above zmax, straight CRUST pass thru
inside of range
calculate target-taper values
*** THIS IS NEW ***
Added an additional backend query
to get the original interp_crust value
and brought it back from backend to use for this
--- only implemented for plugin modules
*** What does this mean?
unless the non-plugin module set these values,
tapering is not going to work. ie. cvmh
can not be tapered, (not sure if it makes sense)
but cvmhlabn could be tapered.
if interp_crust.vs < target-taper.vs
use interp_crust
else
use target-taper
default vs_floor 500, vp_floor 1700, density_floor 1700
if there is a user-defined floor from ucvm_query command line option, use user-defined floors
orig_ratio= vp/vs
Rule 1,
if vs < vs_floor:
vs=vs_floor
vp=vs_floor * orig_ratio
Rule 2,
if vp < vp_floor:
vp=vp_floor
Rule 3,
if density < density_floor:
density=density_floor
Rule 4,
if (vp/vs) < 1.45:
vs=vp/1.45
#define UCVM_DEFAULT_VS_FLOOR 500.0 #define UCVM_DEFAULT_VP_FLOOR 1700.0 #define UCVM_DEFAULT_DENSITY_FLOOR 1700.0
Generate Z1.0 and Z2.5
on Discovery, commands used to create Z1.0 and Z2.5 basin query results,
#!/bin/bash
if [ -z "$UCVM_INSTALL_PATH" ]; then
echo "Need to set UCVM_INSTALL_PATH to run >" ${0##*/}
exit
fi
source $UCVM_INSTALL_PATH/conf/ucvm_env.sh
BIN_DIR=${UCVM_INSTALL_PATH}/bin
CONF_DIR=${UCVM_INSTALL_PATH}/conf
TEST=basin_query_mpi_complete_cvmsi_taper_z2.5
salloc ${UCVM_SALLOC_ENV} -Q --nodes=2 --ntasks=4 --mem=16GB --time=01:00:00 srun -Q -o ${TEST}.srun.out ${BIN_DIR}/basin_query_mpi_complete -b ${TES
T}.first,${TEST}.firstOrSecond,${TEST}.last,${TEST}.secondOnly,${TEST}.threeLast -o ${TEST}.result,${TEST}.meta.json -f ${CONF_DIR}/ucvm.conf -m cvms
i,elygtl:taper -i 10 -v 2500 -l 33.25,-119.38 -s 0.005 -x 578 -y 366
TEST=basin_query_mpi_complete_cvmsi_taper_z1.0
salloc ${UCVM_SALLOC_ENV} -Q --nodes=2 --ntasks=4 --mem=16GB --time=01:00:00 srun -Q -o ${TEST}.srun.out ${BIN_DIR}/basin_query_mpi_complete -b ${TES
T}.first,${TEST}.firstOrSecond,${TEST}.last,${TEST}.secondOnly,${TEST}.threeLast -o ${TEST}.result,${TEST}.meta.json -f ${CONF_DIR}/ucvm.conf -m cvms
i,elygtl:taper -i 10 -v 1000 -l 33.25,-119.38 -s 0.005 -x 578 -y 366
the .firstOrSecond files are the ones that is needed for CyberShake study
Plot the binary file
To plot these files with ucvm_plotting, data ingest mode of plot_z10_map.py and plot_z25_map.py from ucvm_plotting is used but first needs to change those file name to have .binary file name so plotting scripts know how to parse the data
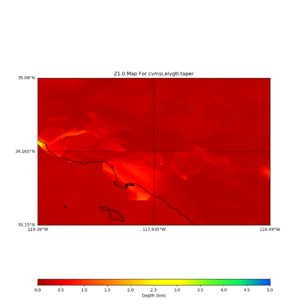
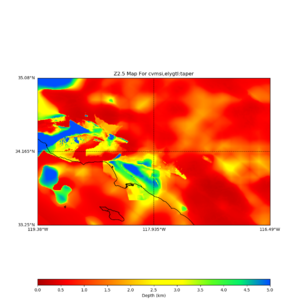
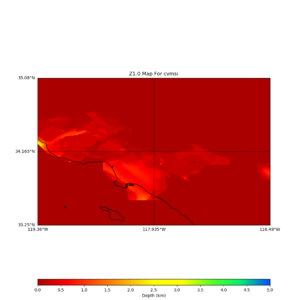
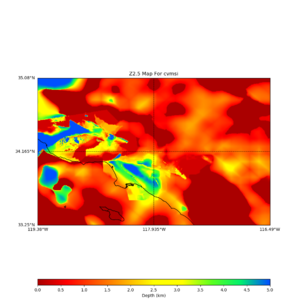
plot_z10_map.py -a s -s 0.0050 -b 33.25,-119.38 -u 35.08,-116.49 -c cvmsi,elygtl:taper -x 578 -y 366 -f cvmsi_taper_z1.0.binary -o cvmsi_taper_z1.0.png plot_z25_map.py -a s -s 0.005 -b 33.25,-119.38 -u 35.08,-116.49 -c cvmsi,elygtl:taper -x 578 -y 366 -f cvmsi_taper_z2.5.binary -o cvmsi_taper_z2.5.png plot_z10_map.py -a s -s 0.0050 -b 33.25,-119.38 -u 35.08,-116.49 -c cvmsi -x 578 -y 366 -f cvmsi_z1.0.binary -o cvmsi_z1.0.png plot_z25_map.py -a s -s 0.005 -b 33.25,-119.38 -u 35.08,-116.49 -c cvmsi -x 578 -y 366 -f cvmsi_z2.5.binary -o cvmsi_z2.5.png
An example of the output,
Using parameters: datafile = cvmsi_taper_z1.0.binary lat1 = 33.25 lat2 = 35.08 color = s outfile = cvmsi_taper_z1.0.png spacing = 0.0050 nx = 578 ny = 366 cvm = cvmsi,elygtl:taper lon1 = -119.38 lon2 = -116.49 Retrieving data. Please wait... TOTAL number of binary data read:211548
Create difference map between 2 binary files
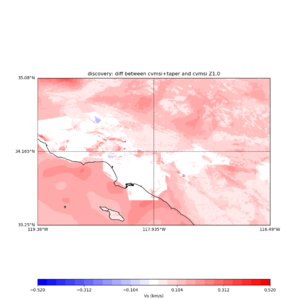
plot_vs30_etree_difference_map.py -s 0.005 -c cvmsi -a d -i ${UCVM_INSTALL_PATH} -b 33.25,-119.38 -u 35.08,-116.49 -o diff_taper_none_z1.0.png -f cvmsi_taper_z1.0.binary,cvmsi_z1.0.binary -x 578 -y 366 -t "diff between cvmsi and cvmsi+taper Z1.0" -A -0.52,0.52
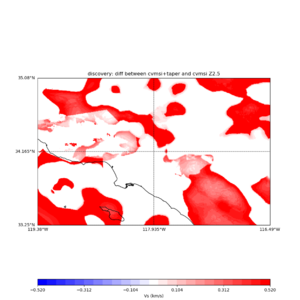
plot_vs30_etree_difference_map.py -s 0.005 -c cvmsi -a d -i ${UCVM_INSTALL_PATH} -b 33.25,-119.38 -u 35.08,-116.49 -o diff_taper_none_z2.5.png -f cvmsi_taper_z2.5.binary,cvmsi_z2.5.binary -x 578 -y 366 -t "diff between cvmsi and cvmsi+taper Z2.5" -A -0.52,0.52
An example of the output :
Using parameters: ny = 366 lat1 = 33.25 datafile2 = cvmsi_z1.0.binary datafile1 = cvmsi_taper_z1.0.binary lat2 = 35.08 color = d title = diff between cvmsi and cvmsi+taper Z1.0 spacing = 0.005 nx = 578 outfile = diff_taper_none_z1.0.png cvm = cvmsi lon1 = -119.38 scalemax = 0.52 scalemin = -0.52 lon2 = -116.49 installdir = /var/www/html/UCVM_web/web/model/UCVM_TARGET Retrieving data. Please wait... Using --> cvmsi_taper_z1.0.binary TOTAL number of binary data read:211548 Using --> cvmsi_z1.0.binary TOTAL number of binary data read:211548
What is actual Z1.0 and Z2.5 for a point
LAPD point -118.125 34.557
#!/bin/sh echo "z2.5 with taper" basin_query -m cvmsi,elygtl:taper -v 2500.0 < in echo "z2.5 no taper" basin_query -m cvmsi -v 2500.0 < in echo "z1.0 with taper" basin_query -m cvmsi,elygtl:taper -v 1000.0 < in echo "1.0 no taper" basin_query -m cvmsi -v 1000.0 < in
using LAPD z2.5 with taper -118.1250 34.5570 580.000 580.000 580.000 z2.5 no taper -118.1250 34.5570 0.000 0.000 0.000 z1.0 with taper -118.1250 34.5570 120.000 120.000 120.000 1.0 no taper -118.1250 34.5570 0.000 0.000 0.000