Difference between revisions of "BBP Batch Scripts"
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | == BBP Batch Scripts == | ||
| + | |||
| + | The following tools were developed to support interactive job submission of suites of bbp scenarios. Currently, on HPCC systems, modified versions of these continue to be used. However, it is rare for users to run them interactively, because this requires waiting long periods of time while the simulations complete. | ||
| + | |||
== Batch Scripts == | == Batch Scripts == | ||
| Line 55: | Line 59: | ||
simulation ID in "./sim_out_dir", if present from a previous run. This would allow the user | simulation ID in "./sim_out_dir", if present from a previous run. This would allow the user | ||
to re-run a simulation with a previously used simulation ID. | to re-run a simulation with a previously used simulation ID. | ||
| + | |||
| + | == Running on USC HPCC == | ||
| + | |||
| + | A sample set of simulations from the Fling study were run on USC HPCC. The original fling generation scripts, source descriptions, station lists, and batch scripts were copied over from broadband.usc.edu to /home/rcf-104. Then small modifications were made to update paths and block the actual execution of the platform (the platform will be run in a PBS job): | ||
| + | |||
| + | Sample scripts can be found at the following locations. However then are not necessarily used in the following order. | ||
| + | |||
| + | {| class="wikitable" | border="1" | ||
| + | |- | ||
| + | ! Script | ||
| + | ! Location | ||
| + | ! Description | ||
| + | ! Modified | ||
| + | |- | ||
| + | | build_xml.py | ||
| + | | /auto/rcf-104/patrices/bbp/batch_tools | ||
| + | | Builds XML workflows for a simulation | ||
| + | | No | ||
| + | |- | ||
| + | | batch_run_bbp.py | ||
| + | | /auto/rcf-104/patrices/bbp/batch_tools | ||
| + | | Executes BBP workflow | ||
| + | | Modified to only write BBP command-lines for simulations to a log for later execution by run_parallel.py. BBP invocations are saved in batch_run_bbp_sims.log and bbp output directory moves are saved in batch_run_bbp_moves.log | ||
| + | |- | ||
| + | | run_parallel.py | ||
| + | | /auto/rcf-104/patrices/bbp/batch_tools | ||
| + | | Helper script to run N programs on a set of M cores | ||
| + | | New script | ||
| + | |- | ||
| + | | gen_source_input.csh | ||
| + | | /auto/rcf-104/patrices/bbp/fling | ||
| + | | Generate full study inputs | ||
| + | | No | ||
| + | |- | ||
| + | | run_bbp-parallel.csh | ||
| + | | /auto/rcf-104/patrices/bbp/fling | ||
| + | | Originally intended to execute the study with the platform. After modifications, only generates XML and execution lists for run_parallel.py. | ||
| + | | Some paths changed, also added ${ROOT_PATH} to some relative path locations to make them absolute paths | ||
| + | |} | ||
| + | |||
| + | |||
| + | General steps for running the Fling study: | ||
| + | |||
| + | * Generate inputs | ||
| + | <pre> | ||
| + | $ ./gen_source_input.csh | ||
| + | </pre> | ||
| + | * Generate XML workflows | ||
| + | <pre> | ||
| + | $ ./run_bbp-parallel.csh | ||
| + | </pre> | ||
| + | * Create PBS job submission script (example below) | ||
| + | * Submit PBS job to USC HPCC | ||
| + | |||
| + | |||
| + | Example PBS script running the sample Fling simulations on 16 cores: | ||
| + | |||
| + | <pre> | ||
| + | #!/bin/bash | ||
| + | |||
| + | #PBS -q nbns | ||
| + | #PBS -l arch=x86_64,pmem=2000mb,pvmem=3000mb,walltime=6:00:00,nodes=4:ppn=4 | ||
| + | #PBS -V | ||
| + | #PBS -e /home/rcf-104/patrices/bbp/fling/Xml1/Set1/run_set1.err | ||
| + | #PBS -o /home/rcf-104/patrices/bbp/fling/Xml1/Set1/run_set1.out | ||
| + | |||
| + | PYTHONPATH=/home/rcf-104/patrices/bbp/11.2.2/bbp_2g/comps | ||
| + | |||
| + | HOME=/home/rcf-104/patrices/bbp/fling | ||
| + | |||
| + | echo "Jobs start" | ||
| + | date | ||
| + | |||
| + | cd $HOME | ||
| + | |||
| + | python $HOME/Xml1/Set1/run_parallel.py /home/rcf-104/patrices/bbp/11.2.2/setup_bbp_env.sh $HOME | ||
| + | /Xml1/Set1/batch_run_bbp_sims.log $PBS_NODEFILE 1 | ||
| + | |||
| + | python $HOME/Xml1/Set1/run_parallel.py /home/rcf-104/patrices/bbp/11.2.2/setup_bbp_env.sh $HOME | ||
| + | /Xml1/Set1/batch_run_bbp_moves.log $PBS_NODEFILE 1 | ||
| + | |||
| + | echo "Jobs end" | ||
| + | date | ||
| + | </pre> | ||
| + | |||
| + | == Comparison of Seismogram from Server and Cluster == | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! Simulation | ||
| + | ! broadband.usc.edu | ||
| + | ! USC HPCC cluster | ||
| + | |- | ||
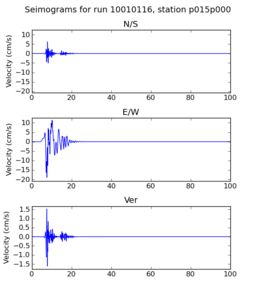
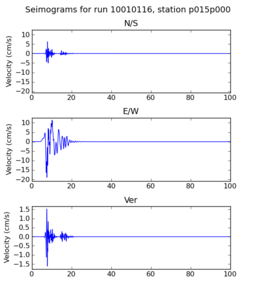
| + | | 10010116 | ||
| + | | [[File: 10010116_ref.p015p000_velocity_seis.png|256px|thumb]] | ||
| + | | [[File: 10010116_hpcc.p015p000_velocity_seis.png|256px|thumb]] | ||
| + | |- | ||
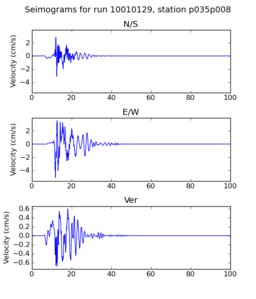
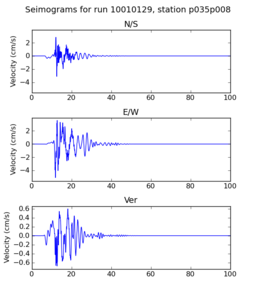
| + | | 10010129 | ||
| + | | [[File: 10010129_ref.p035p008_velocity_seis.png|256px|thumb]] | ||
| + | | [[File: 10010129_hpcc.p035p008_velocity_seis.png|256px|thumb]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | == Certification of USC HPCC Cluster for Broadband Calculations == | ||
| + | |||
| + | The verification and validation of the currently released Broadband platform is based on results generated on a SCEC server called broadband.usc.edu. When we move the Broadband platform software, re-build it, and re-run it in a different computing environment, the results the platform produces can be slightly different than results produced on the SCEC server. Differences can come from computing hardware, from operating system characteristics, from compiler version, and other sources. | ||
| + | |||
| + | Before accepting results generated in a new computing environment, we must first certify that the new computing environment produces results that are equivalent to the results from the original server where the platform was originally developed and tested. | ||
| + | |||
| + | To speed up execution of the Fling study, we plan to run it on the USC HPCC cluster, so we must certify that USC HPCC cluster results are valid and comparable to those generated on broadband.usc.edu. | ||
| + | |||
| + | Below are initial results from our initial certification tests. A researcher ran a small subset of the Fling study on the SCEC broadband server. Then, we ran the same subset on the USC HPCC cluster. Below we compare the output seismograms from both runs, showing that the two results are very similar. | ||
| + | |||
| + | In our discussions, we decided that the certification criteria for this study will include a number of small magnitude ruptures, and a number of large magnitude ruptures, which we will post when they are available. | ||
| + | |||
| + | == Building Metrics Table == | ||
| + | The following command will generate the metrics table above. | ||
| + | |||
| + | <pre> | ||
| + | $ tot=0; for i in `ls | grep "Scenario"`; do echo -n "$i "; cnt=`cat $i/StatInfo/*.stl | grep -v "#" | wc -l` ; echo -n "$cnt "; \ | ||
| + | tot=$(($tot+$cnt)); num_smgr=$(($cnt*30)); echo $num_smgr; done; echo "$tot $(($tot*30))" | ||
| + | </pre> | ||
Latest revision as of 00:27, 16 May 2012
Contents
BBP Batch Scripts
The following tools were developed to support interactive job submission of suites of bbp scenarios. Currently, on HPCC systems, modified versions of these continue to be used. However, it is rare for users to run them interactively, because this requires waiting long periods of time while the simulations complete.
Batch Scripts
BBP_2G Batch Automation Tools v1.2, 2011-08-19
This release includes two Python scripts for batch automation of Broadband Platform simulations.
build_xml.py:
Script to generate XML files formatted for BBP_2G. The XML files are created based on user provided run description files. This script takes a path to the folder containing run description files as input and generates a set of XML files. These XML files can be used as inputs to automate the run of Broadband simulations.
Usage example: build_xml.py -i "/home/user/run_descs" -x "/home/user/run_descs"
This will parse the run description files in "/home/user/run_descs" and generate XML files with BBP and save them in "/home/user/run_descs" folder.
This script expects run description text files with the following format: RUN_TAG = 10010100 VALIDATION_RUN = n SOURCE_DESCRIPTION_FILE = /home/NgaW2/FwHw/FaultInfo/Inputs/m6.00_d20_r90_z0.src STATION_LIST_FILE = /home/NgaW2/FwHw/StatInfo/rv01-m6.00_stats.stl RUPTURE_GENERATOR = URS LOW_FREQUENCY_MODULE = URS HIGH_FREQUENCY_MODULE = URS SITE_RESPONSE_MODULE = URS PLOT_VEL = y PLOT_ACC = y RUN_GOF = n
Note: RUN_TAG is equivalent to simulation ID
batch_run_bbp.py:
Script to run a set of Broadband simulation based on input XML files. This script takes a path to a folder containing BBP formated XML files and runs simulations with BBP for each of the XML files in the specified folder. The script has a build in resume feature which allows the script to track and skip XML files which were previously processed. An output directory can be specified to collate simulation directories (indata, outdata tmpdata and logs) in one location.
Usage example: batch_run_bbp.py -i "./run_xml" -o "./sim_out_dir" -r -f
This will run the Broadband Platform with each of the XML files in "./run_xml" folder. The -o option will cause the script to move simulation directories (indata/<simID>, outdata/<simID>, tmpdata/simID, logs/simID) to "./sim_out_dir" folder. The -r option will allow the script to skip all the XML files that were previously processed. The -f option will allow the script to force overwrite any BBP folders indata/<simID>, outdata/<simID>, tmpdata/<simID>, logs/<simID>. The script will also overwrite simulation folders with same simulation ID in "./sim_out_dir", if present from a previous run. This would allow the user to re-run a simulation with a previously used simulation ID.
Running on USC HPCC
A sample set of simulations from the Fling study were run on USC HPCC. The original fling generation scripts, source descriptions, station lists, and batch scripts were copied over from broadband.usc.edu to /home/rcf-104. Then small modifications were made to update paths and block the actual execution of the platform (the platform will be run in a PBS job):
Sample scripts can be found at the following locations. However then are not necessarily used in the following order.
| Script | Location | Description | Modified |
|---|---|---|---|
| build_xml.py | /auto/rcf-104/patrices/bbp/batch_tools | Builds XML workflows for a simulation | No |
| batch_run_bbp.py | /auto/rcf-104/patrices/bbp/batch_tools | Executes BBP workflow | Modified to only write BBP command-lines for simulations to a log for later execution by run_parallel.py. BBP invocations are saved in batch_run_bbp_sims.log and bbp output directory moves are saved in batch_run_bbp_moves.log |
| run_parallel.py | /auto/rcf-104/patrices/bbp/batch_tools | Helper script to run N programs on a set of M cores | New script |
| gen_source_input.csh | /auto/rcf-104/patrices/bbp/fling | Generate full study inputs | No |
| run_bbp-parallel.csh | /auto/rcf-104/patrices/bbp/fling | Originally intended to execute the study with the platform. After modifications, only generates XML and execution lists for run_parallel.py. | Some paths changed, also added ${ROOT_PATH} to some relative path locations to make them absolute paths |
General steps for running the Fling study:
- Generate inputs
$ ./gen_source_input.csh
- Generate XML workflows
$ ./run_bbp-parallel.csh
- Create PBS job submission script (example below)
- Submit PBS job to USC HPCC
Example PBS script running the sample Fling simulations on 16 cores:
#!/bin/bash #PBS -q nbns #PBS -l arch=x86_64,pmem=2000mb,pvmem=3000mb,walltime=6:00:00,nodes=4:ppn=4 #PBS -V #PBS -e /home/rcf-104/patrices/bbp/fling/Xml1/Set1/run_set1.err #PBS -o /home/rcf-104/patrices/bbp/fling/Xml1/Set1/run_set1.out PYTHONPATH=/home/rcf-104/patrices/bbp/11.2.2/bbp_2g/comps HOME=/home/rcf-104/patrices/bbp/fling echo "Jobs start" date cd $HOME python $HOME/Xml1/Set1/run_parallel.py /home/rcf-104/patrices/bbp/11.2.2/setup_bbp_env.sh $HOME /Xml1/Set1/batch_run_bbp_sims.log $PBS_NODEFILE 1 python $HOME/Xml1/Set1/run_parallel.py /home/rcf-104/patrices/bbp/11.2.2/setup_bbp_env.sh $HOME /Xml1/Set1/batch_run_bbp_moves.log $PBS_NODEFILE 1 echo "Jobs end" date
Comparison of Seismogram from Server and Cluster
| Simulation | broadband.usc.edu | USC HPCC cluster |
|---|---|---|
| 10010116 | ||
| 10010129 |
Certification of USC HPCC Cluster for Broadband Calculations
The verification and validation of the currently released Broadband platform is based on results generated on a SCEC server called broadband.usc.edu. When we move the Broadband platform software, re-build it, and re-run it in a different computing environment, the results the platform produces can be slightly different than results produced on the SCEC server. Differences can come from computing hardware, from operating system characteristics, from compiler version, and other sources.
Before accepting results generated in a new computing environment, we must first certify that the new computing environment produces results that are equivalent to the results from the original server where the platform was originally developed and tested.
To speed up execution of the Fling study, we plan to run it on the USC HPCC cluster, so we must certify that USC HPCC cluster results are valid and comparable to those generated on broadband.usc.edu.
Below are initial results from our initial certification tests. A researcher ran a small subset of the Fling study on the SCEC broadband server. Then, we ran the same subset on the USC HPCC cluster. Below we compare the output seismograms from both runs, showing that the two results are very similar.
In our discussions, we decided that the certification criteria for this study will include a number of small magnitude ruptures, and a number of large magnitude ruptures, which we will post when they are available.
Building Metrics Table
The following command will generate the metrics table above.
$ tot=0; for i in `ls | grep "Scenario"`; do echo -n "$i "; cnt=`cat $i/StatInfo/*.stl | grep -v "#" | wc -l` ; echo -n "$cnt "; \ tot=$(($tot+$cnt)); num_smgr=$(($cnt*30)); echo $num_smgr; done; echo "$tot $(($tot*30))"